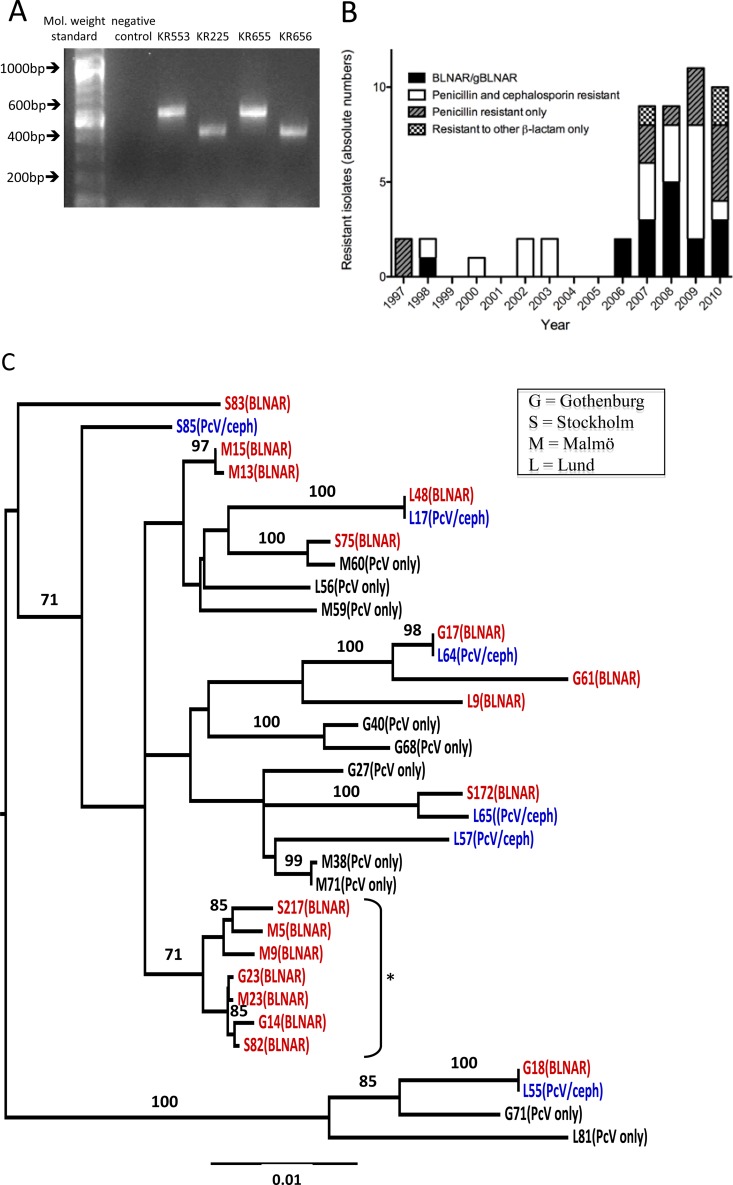
Fig 2.
Two variants of blaTEM-1 and a steep increase of β-lactamase-negative invasive isolates with a cluster of BLNAR isolates were identified. (A) The agarose gel shows an example of a blaTEM-1 PCR result from four different invasive NTHi strains with β-lactamase production. The lanes are, from left to right, molecular weight standard, negative control, and the clinical NTHi isolates KR553, KR225, KR655, and KR656. Sequencing revealed that the products of KR553 and KR655 are blaTEM-1 wild type, whereas the products of KR225 and KR656 are representative of the blaTEM-1-PΔ. (B) The recent increase of H. influenzae isolates with a β-lactamase-negative, β-lactam-resistant phenotype. The absolute numbers of invasive BLNBR isolates during 1997 to 2010, sorted by resistance phenotype, are shown. The black bars show BLNAR isolates, and the white bars show isolates resistant to penicillin and a cephalosporin. The striped gray bars show isolates resistant to penicillin only, while the checked bars show isolates resistant to only a cephalosporin or a carbapenem. (C) A neighbor-joining phylogenetic tree was constructed based on concatenated MLST data from all available invasive BLNBR isolates. The BLNAR isolates are indicated in red. Isolates with penicillin and cephalosporin (PcV/ceph) resistance are indicated in blue, while isolates with only penicillin (PcV only) resistance are shown in black. The prefix letter of the isolate name indicates the laboratory where the isolate was isolated: G, Gothenburg; S, Stockholm; M, Malmö; L, Lund. Clusters with >70% bootstrap support are indicated with their bootstrap values, and one cluster of seven gBLNAR/BLNAR isolates, including isolates from all three geographical areas of the study, is indicated by an asterisk.