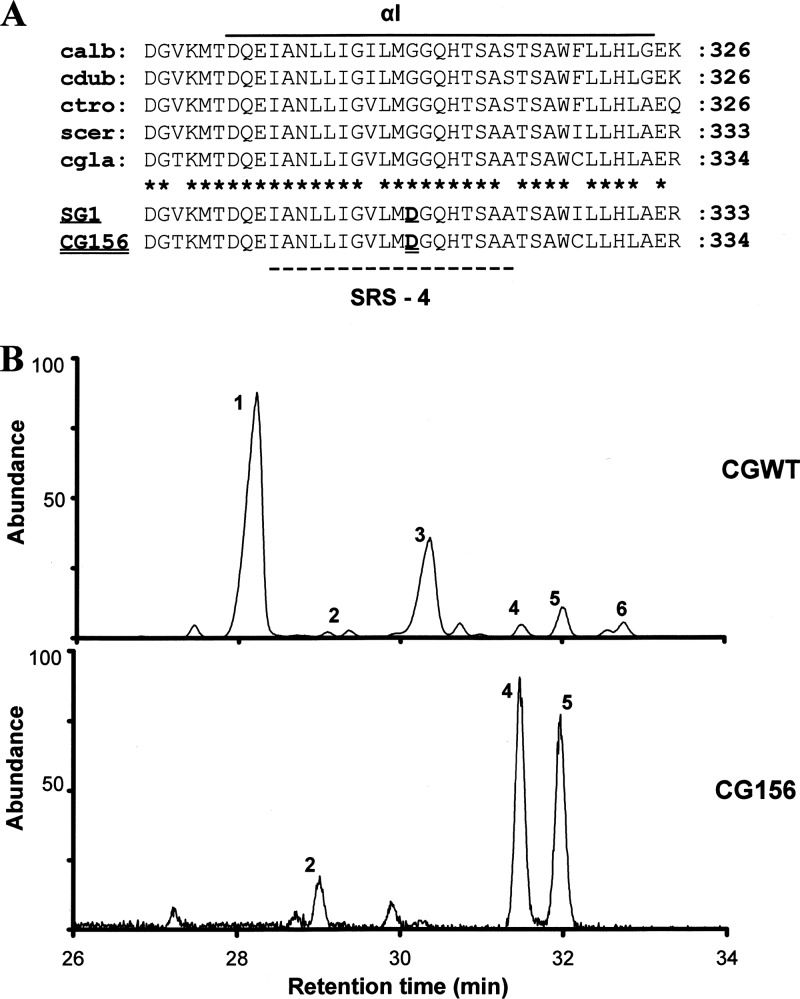
Fig 3.
ERG11 protein sequence analysis and complementation studies. (A) Alignment of sterol 14α-demethylase sequences (αI helix only) for Candida albicans (calb; accession number P10613), C. dubliniensis (cdub; AAK57519), C. tropicalis (ctro; P14263), Saccharomyces cerevisiae (scer; AAA34379), and C. glabrata (cgla; P50859). SRS-4, substrate recognition site 4. Asterisks indicate conserved residues. Note that a glycine-to-aspartate alteration previously reported to nullify the function of sterol 14α-demethylase in SG1, an S. cerevisiae erg11 mutant (13), was the sole amino acid substitution detected in CG156 (this study). (B) Sterol chromatograms for S. cerevisiae constructs heterologously expressing C. glabrata wild-type (CGWT) and mutated (CG156) sterol 14α-demethylase. Peak 1, ergosterol; 2, 4,14α-dimethyl zymosterol; 3, 4-methyl zymosterol; 4,14α-methyl ergosta-8,24(28)-dien-3β,6α-diol; 5, lanosterol; and 6, 4,4-dimethyl zymosterol.