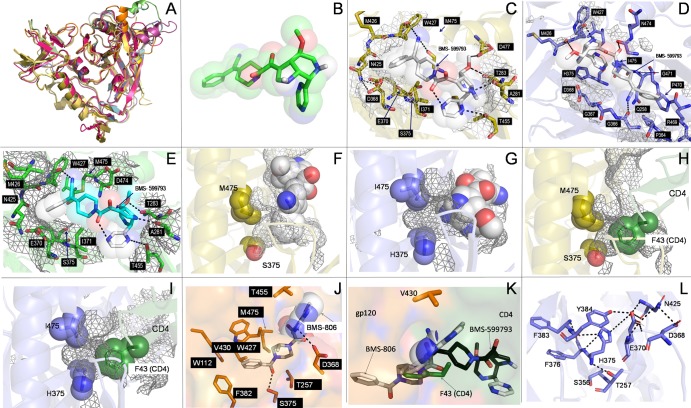
Fig 6.
In silico modeling and docking simulations. Alignment of multiple gp120 models and structures showed excellent global structural similarities (A). Three-dimensional structure of BMS-599793 (B). Close-ups of BMS-599793 (white) docked to subtype B gp120 (PDB ID:2B4C) (C), CRF01_AE gp120 (PDB ID: 3SE8) (D), and modeled subtype B HIV-1HXB2 gp120 (E); interaction face on gp120 is covered with mesh and residues involved in interactions are shown as sticks (C, D, and E). The relative positions of residues 375 and 475 to BMS-599793 (white) in subtype B gp120 (F) (orange) and CRF01_AE gp120 (G) (blue); mesh represents internal cavities in gp120 (F and G). The relative positions of residues 375 and 475 to F-43 of superimposed CD4 in subtype B gp120 (H) and CRF01_AE gp120 (I). Close-up of BMS-806 docked to subtype B gp120 (J); key residues in interaction are shown in stick form; buried portion of BMS-806 is shown only as sticks while solvent-exposed portion is shown as sticks within partially transparent space-filling structure. Superimposition of BMS-806 (white), BMS-599793 (black), and CD4 F-43 (green) showing their relative depths of penetration into the F-43 cavity of subtype-B gp120 (K). Hydrophilic and electrostatic interactions that potentially disrupt and prevent insertion of BMS-599793 into the F43 cavity of CRF01_AE gp120 (L). All dotted lines in the figure refer to postulated hydrogen bonding distances ≥ 3Å or postulated salt-bridge interaction distances ≥ 4Å.