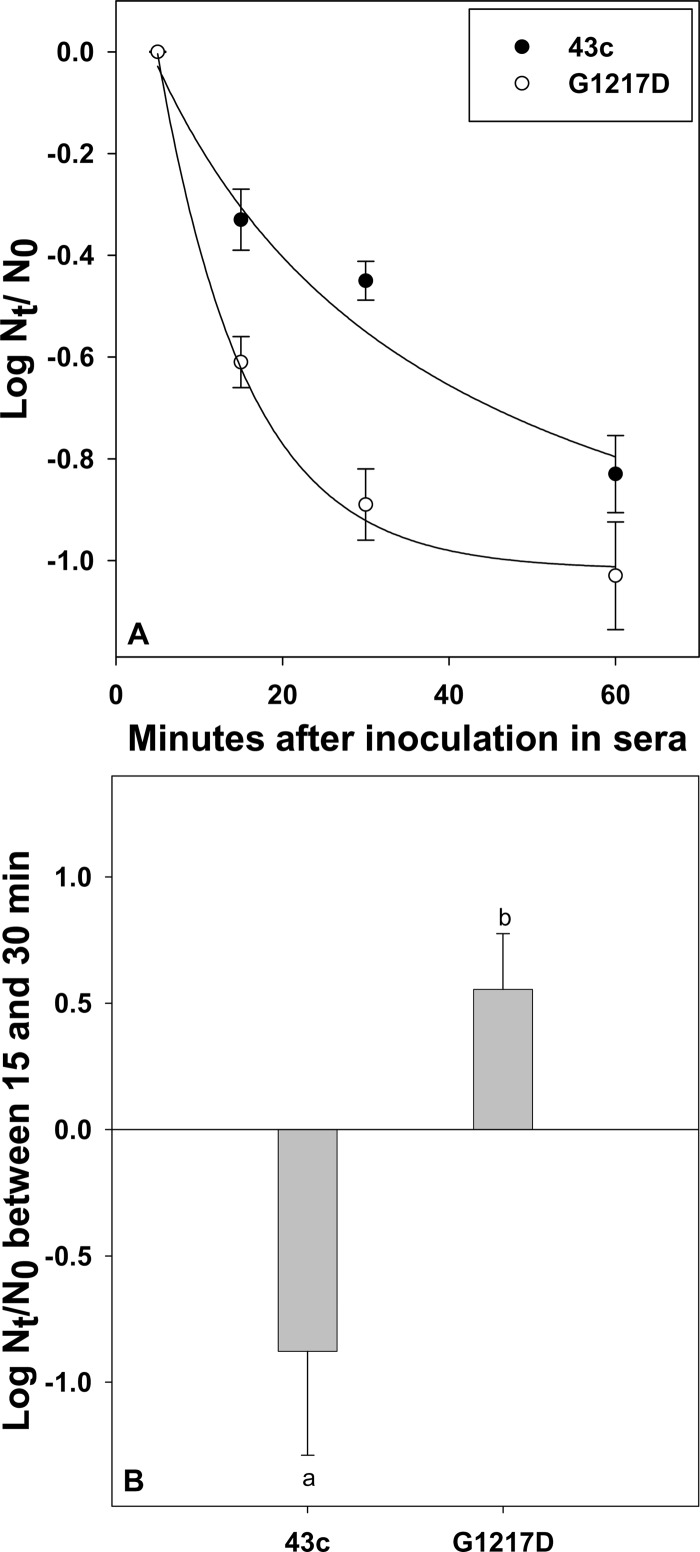
Fig 2.
Comparative kinetics of clearance from blood of HAV parental and G1217D mutant strains. Rats were injected intravenously with 1 ml of virus at 108 genome copies/ml via the tail vein. Blood (200 to 300 μl) was obtained through the saphenous vein at 1, 5, 15, 30, and 60 min p.i., and virus genome copy numbers in both blood compartments (serum and blood cells) were determined. (A) Genome copy number decay in serum, expressed as the log10(Nt/N0) value, where Nt is the genome copy number at 15, 30, or 60 min p.i. and N0 is the genome copy number at 5 min p.i. Curves were fitted by nonlinear regression with SigmaPlot (SPSS, Chicago, IL). (B) Genome copy number variation in blood cells between 15 and 30 min p.i., expressed as the log10(Nt/N0), where Nt is the genome copy number at 30 min p.i. and N0 is the genome copy number at 15 min p.i. Different letters indicate statistically significant differences (P < 0.05).