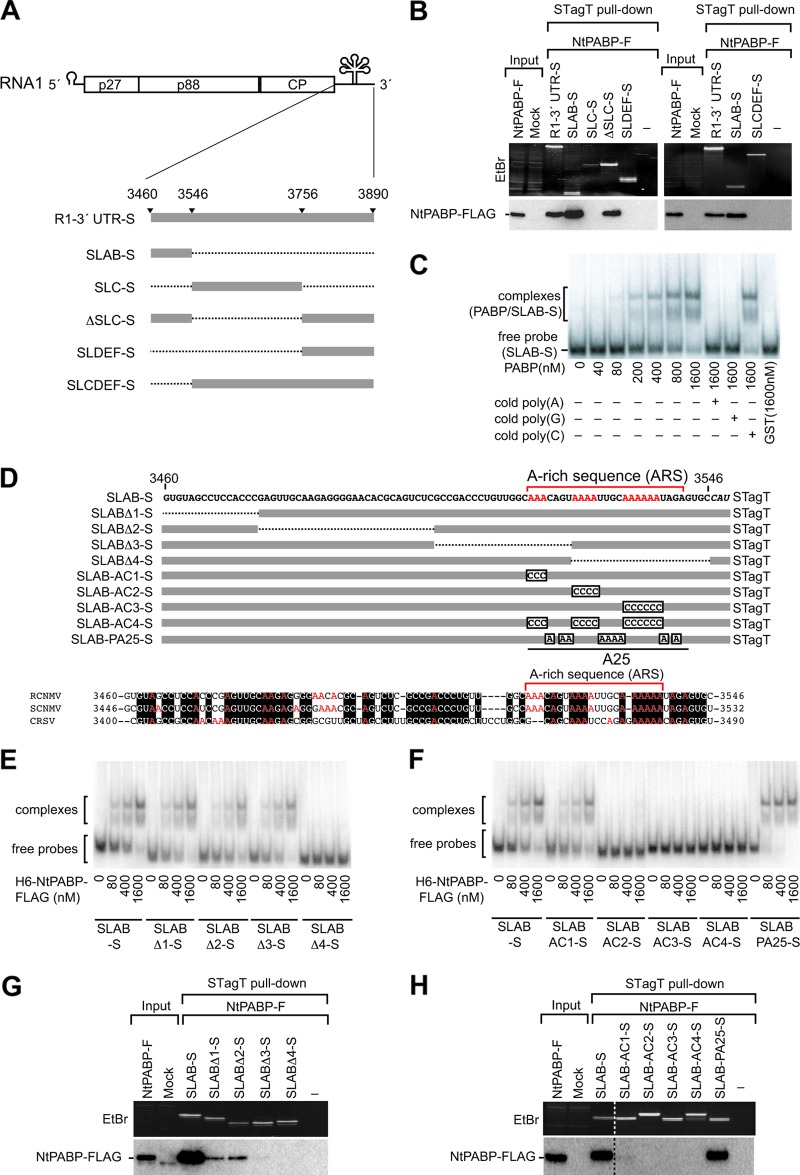
Fig 2.
NtPABP binds specifically to an internal A-rich sequence in the 3′ UTR of RNA1. (A) Schematic representation of STagT-fused viral RNA fragments. Bold lines indicate virus-derived sequences with the nucleotide numbers of RNA1 at the 5′ and 3′ ends. Dashed lines indicate deleted sequence. (B) STagT pulldown assay using truncated mutants of the 3′ UTR of RNA1 in BYLS20 expressing FLAG-tagged NtPABP. (C) EMSA for NtPABP with SLAB region. The specificity of the NtPABP-SLAB interaction was determined by adding RNA competitors [poly(A), poly(G), and poly(C); Sigma-Aldrich]. (D) At the top is a schematic representation of STagT-fused viral RNA fragments. Bold lines indicate virus-derived sequences. Dashed lines indicate deleted sequences. Substituted sequences are shown in boxes. At the bottom, alignments of the SLAB region of RNA1 in dianthoviruses including RCNMV, Sweet clover necrotic mosaic virus (SCNMV), and Carnation ring spot virus (CRSV) are shown. Conserved nucleotides are highlighted with black boxes and white letters. Adenine residues are highlighted with red letters. (E and F) EMSA for NtPABP using SLAB and its mutants. (G and H) STagT pulldown assay using SLAB and its mutants in BYLS20 expressing FLAG-tagged NtPABP.