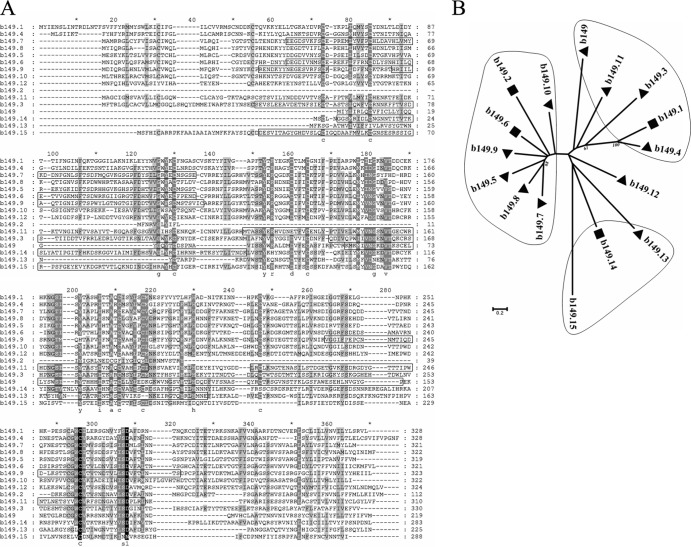
Fig 5.
Analysis of the MsHV b149 family. (A) Alignment of the b149 family using ClustalW. The sequences in boxes were predicted to fold like immunoglobulin-like beta-sandwich domains by the software program Phyre2. (B) Neighbor-joining tree of the b149 family, based on the sequence alignment. The tree was reconstructed using MEGA5, with the parameter setting of the Jones-Taylor-Thornton model, gaps/missing-data treatment of pairwise deletion, and 1,000 bootstraps. Bootstrap percentages of more than 50% are shown. Triangles indicate that both the signal peptide at the N terminus and the transmembrane domain at the C terminus were predicted, while rectangles indicate that only the transmembrane domain at the C terminus was predicted.