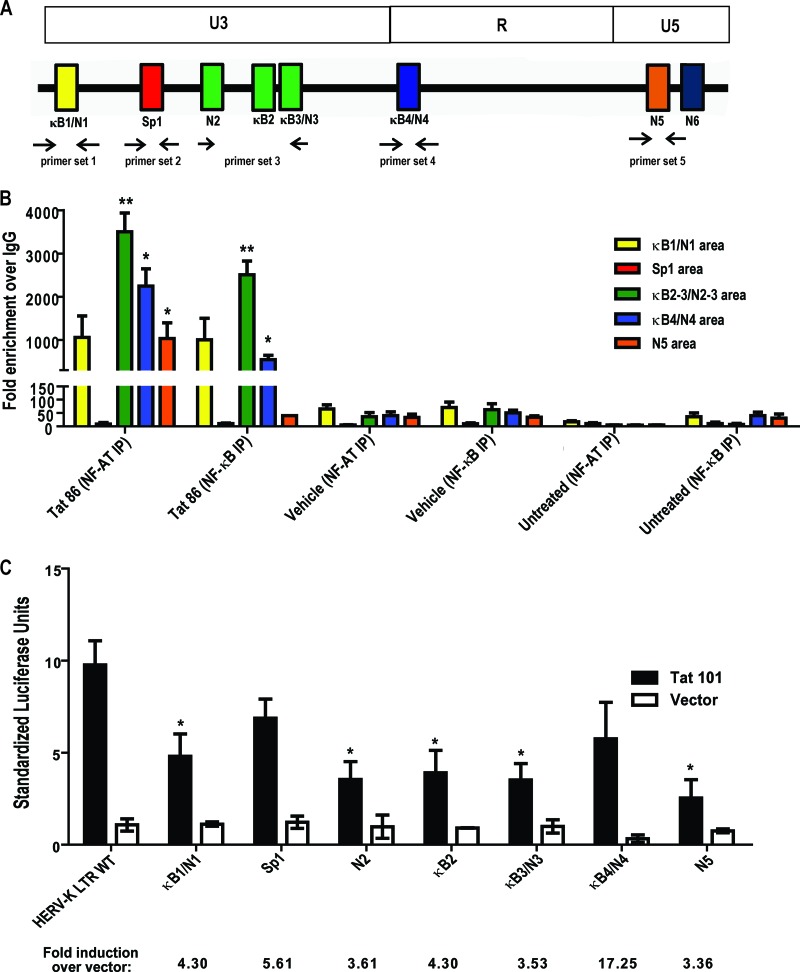
Fig 6.
Tat responsive elements in the HERV-K (HML-2) promoter. ChIP was performed on the cellular HERV-K (HML-2) LTR promoter in Jurkat T cells using antibodies specific for NF-κB or NF-AT (or nonspecific IgG isotype controls) 1 h after treatment with either recombinant HIV-1 Tat protein or PMA and ionomycin. qPCR was performed using primer sets designed to target areas of the promoter where the potential transcription factor binding sites are present. (A) Schematic of primer binding areas in the HERV-K (HML-2) promoter. (B) ChIP analysis of potential NF-κB and NF-AT binding sites with data expressed as the fold enrichment over IgG. Bars are color coded to match primer binding areas. Error bars indicate the SD from three independent immunoprecipitation experiments. (C) Potential transcription factor binding site mutations in the HERV-K (HML-2) LTR and their response to HIV-1 Tat in Jurkat T cells. WT, wild type; κB, NF-κB; N, NF-AT. Luciferase activation was measured 48 h after transfection. Control experiments included a mock transfection and transfection of an empty vector (pcDNA3.1). Error bars indicate the standard errors of the mean from three independent transfections. Significance was calculated using a Student t test comparing Tat treatments to vehicle controls or full-length LTR activity to mutant-LTR activity. Significant results are indicated (*, P < 0.05; **, P < 0.005).