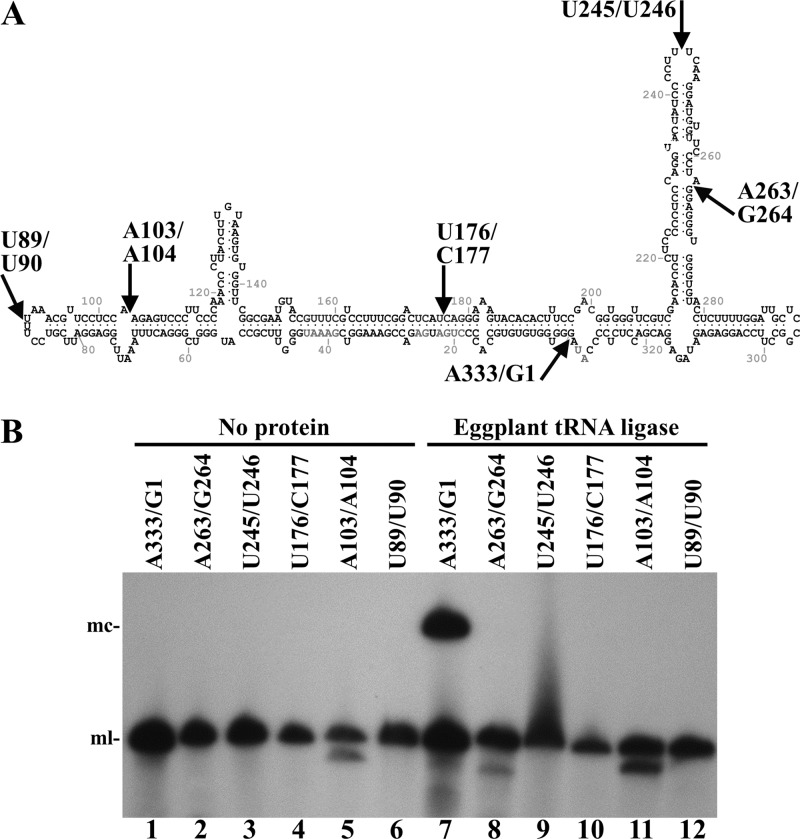
Fig 3.
Circularization by eggplant tRNA ligase of different monomeric linear ELVd (+) RNAs opened at different sites. (A) ELVd folded in the predicted minimum free-energy conformation. Arrows indicate positions in which the different linear ELVd substrates subjected to ligation were opened. (B) ELVd circularization assay with recombinant eggplant tRNA ligase. Reaction products were separated by denaturing PAGE and ELVd (+) RNAs revealed by Northern blot hybridization. Lanes 1 to 6, controls with no protein added. Lanes 7 to 12, eggplant tRNA ligase added. Substrate RNAs were opened between positions A333 and G1 (lanes 1 and 7), A263 and G264 (lanes 2 and 8), U245 and U246 (lanes 3 and 9), U176 and C177 (lanes 4 and 10), A103 and A104 (lanes 5 and 11), and U89 and U90 (lanes 6 and 12). The positions of monomeric circular (mc) and linear (ml) ELVd forms are indicated on the left.