Fig 8.
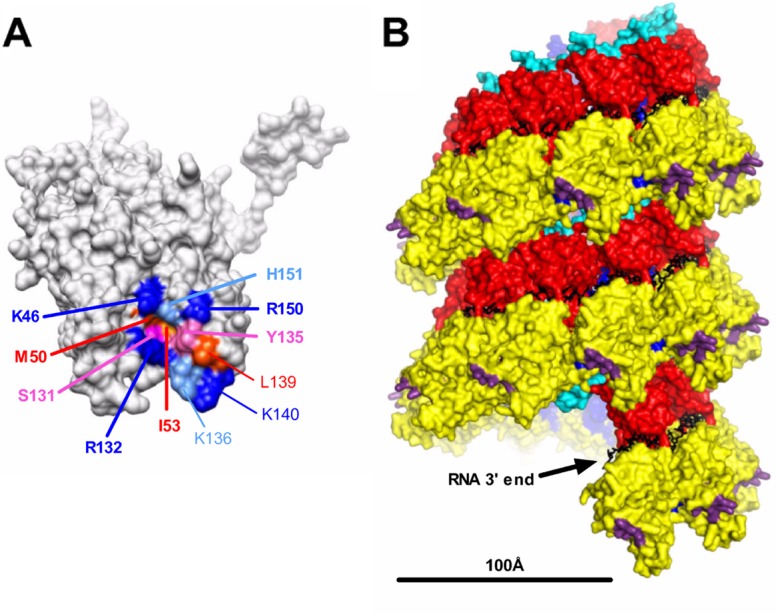
Localization of the PI region on the nucleocapsid. (A) Localization at the surface of the N monomer of residues identified in this study as being critical for RNA synthesis. Residues of PI are indicated in boldface type. Residues are colored according to their biochemical properties (blue, basic residues; red, hydrophobic residues; pink, neutral residues). (B) Side view of the helical nucleocapsid modeled from the ring using a pitch of 70 Å according data reported previously by Tawar et al. (44). Structural domains are colored; i.e., the NCTD and NNTD are in red and yellow, respectively, and the C-arm and N-arm are in light blue and blue, respectively. Residues critical for the interaction with the PCTD are shown in purple, and RNA is shown in black. The bar indicates 100 Å. The structure of the RSV N protein bound to RNA (44) was obtained from the Protein Data Bank (PDB accession number 2WJ8) and visualized by using UCSF Chimera molecular graphics software (34).
