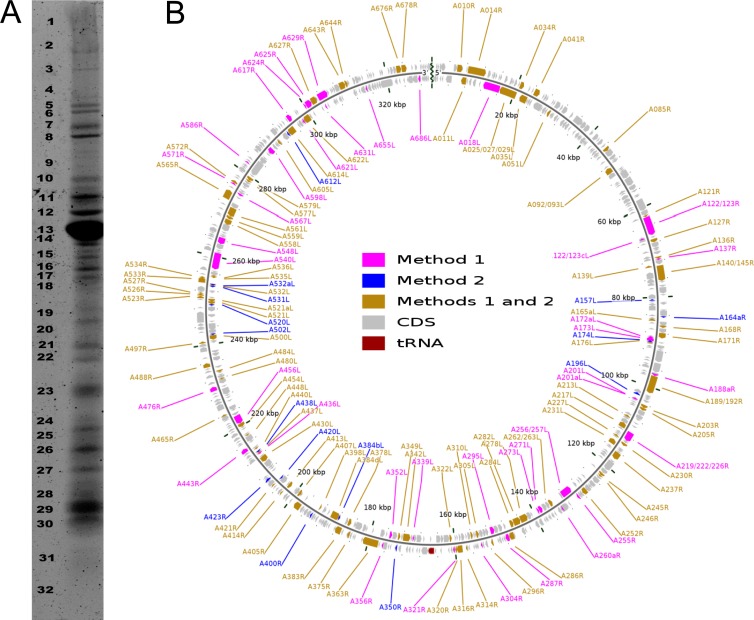
Fig 2.
SDS-PAGE protein separation and virion proteome mapping onto the PBCV-1 genome. (A) Distribution of virion proteins with SDS polyacrylamide gel separation. The numbers on the left indicate the gel fragment that was analyzed. (B) The PBCV-1 genome was resequenced, assembled, and annotated to correct existing sequence errors. The 416 predicted CDSs are represented as gray arrows running both clockwise and counterclockwise along the genome. Note that the diagram is circular, but there is a break at the 12 o'clock position because the viral genome is a linear molecule with terminal inverted repeats and closed hairpin ends. The terminal sequences (inverted repeats and hairpin ends) were found to be identical to those reported previously (67). The polycistronic gene encoding 11 tRNAs is presented in red (at 6 o'clock). The 148 proteins of the virion proteome were determined using two independent mass spectrometry-based methods (see Materials and Methods). The results of each method are shown. The map was developed with CGView software (43).