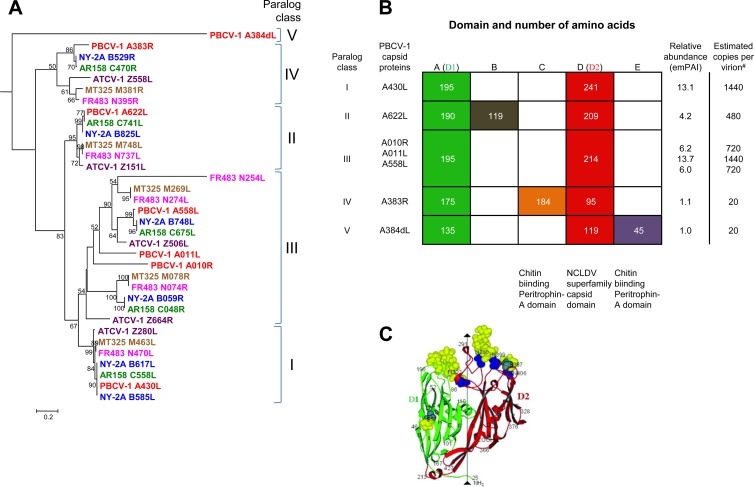
Fig 5.
Capsid protein paralog classes and relative abundances in PBCV-1. (A) The seven capsid-like proteins detected in the PBCV-1 virion were evaluated against a data set of chloroviruses, including PBCV-1 (RefSeq NC_000852.5), NY-2A (RefSeq NC_009898.1), AR158 (RefSeq NC_009899.1), MT325 (GenBank DQ491001.1), FR483 (RefSeq NC_008603.1), and ATCV-1 (RefSeq NC_008724.1). These 7 proteins had homologs in each of the viruses that separated into 5 distinct paralog classes (I to V), as shown in the neighbor-joining tree (see Table S3 in the supplemental material for CDS accession numbers). The sequence for PBCV-1 A384dL, a member of paralog class V, which is distantly related, was used as the outgroup to root the phylogenetic analysis using the website http://www.phylogeny.fr (10). Muscle was used to align the sequences. Bootstrap analysis was used to construct the tree. Similar tree topologies were produced by maximum-likelihood and maximum-parsimony analyses. The values on the branches are the percentages of bootstrap support (200 replicates). Only bootstrap values of >50% are shown. The distance bar represents 0.2 amino acid substitution per site. (B) PBCV-1 capsid proteins grouped into 5 paralog classes within their two conserved domains. The D1 domain (column A) and the D2 NCLDV superfamily capsid domain (column D) were previously determined by structure analysis of the Vp54 MCP (30) (shown in panel C). The relative abundances, as determined by the emPAI method for the method 1 data, are listed on the right, along with the hypothetical estimated number of copies of each capsid protein per virion. Note that the two proteins at relatively low abundance contain chitin binding peritrophin A conserved domains (columns C and E). Column B is a domain with function.