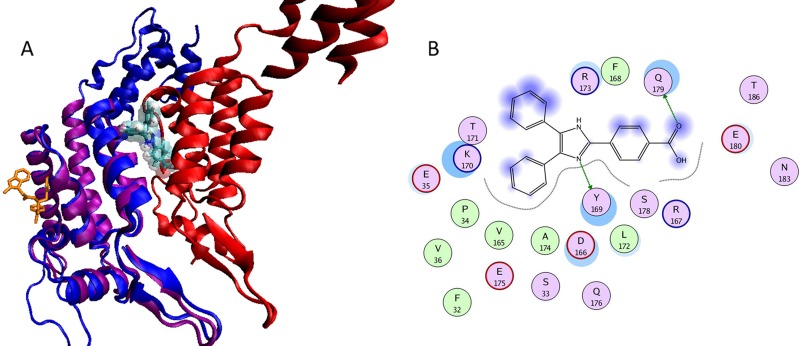
Fig 7.
(A) Comparison of the proposed binding site of I-XW-053 with the binding site of compound PF74. Structural superpositioning of cocrystallized PF74 with NTD of CA protein on CA dimers. The protein is represented in a cartoon model, and the protomers are colored purple, green, and red. PF74 is represented in an orange licorice model, and I-XW-053 is docked at the proposed binding site and is represented in a licorice model and colored by atom type (C in cyan, N in blue, O in red, and H in white). The two binding sites are distinct and opposite to each other. The van der Waals surface model of I-XW-053 clearly shows that I-XW-053 sterically clashes with one of the CA protomers (in red) and hence blocks the assembly of the CA protein. (B) I-XW-053 in the binding site of CA monomer. The binding site residues are colored by their nature, with hydrophobic residues in green, polar residues in purple, and charged residues highlighted with bold contours. Blue spheres and contours indicate matching regions between ligand and receptors. Hydrogen-bonded interactions are shown by green arrows. The figure was generated using the MOE ligX module.