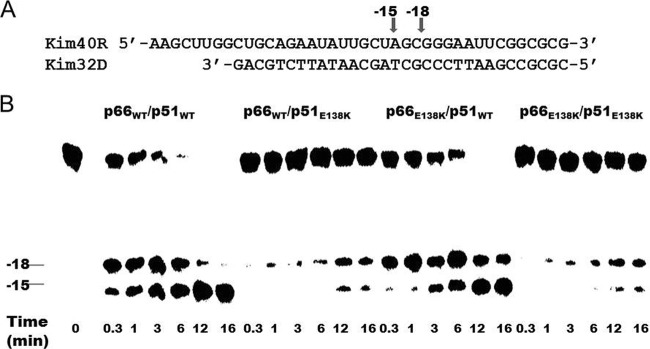
Fig 2.
Subunit-specific analysis of the effect of the E138K mutation in HIV-1 RT on RNase H activity. (A) Graphic representation of the substrate RNA-DNA (kim40R/kim32D) duplex used to monitor the cleavage efficiency of recombinant RT. The 40-mer RNA kim40R was labeled at its 5′ terminus with 32P and annealed to a 32-mer DNA oligonucleotide, kim32D. The positions of cleaved products are indicated by arrows. (B) RNase H activities were analyzed by monitoring substrate cleavage in time course experiments in the presence of a heparin trap. The positions of cleaved products are indicated on the left. The uncleaved substrate indicates the variability in RNase H activity among different RT enzymes. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis.