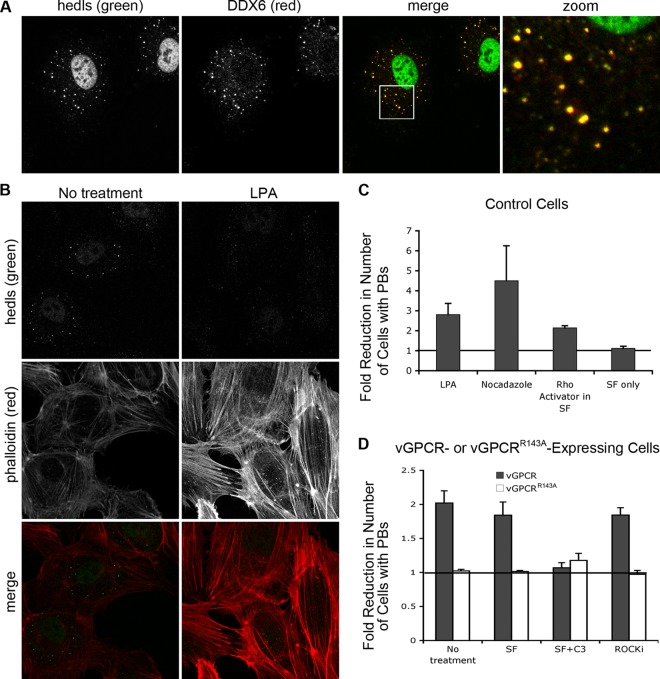
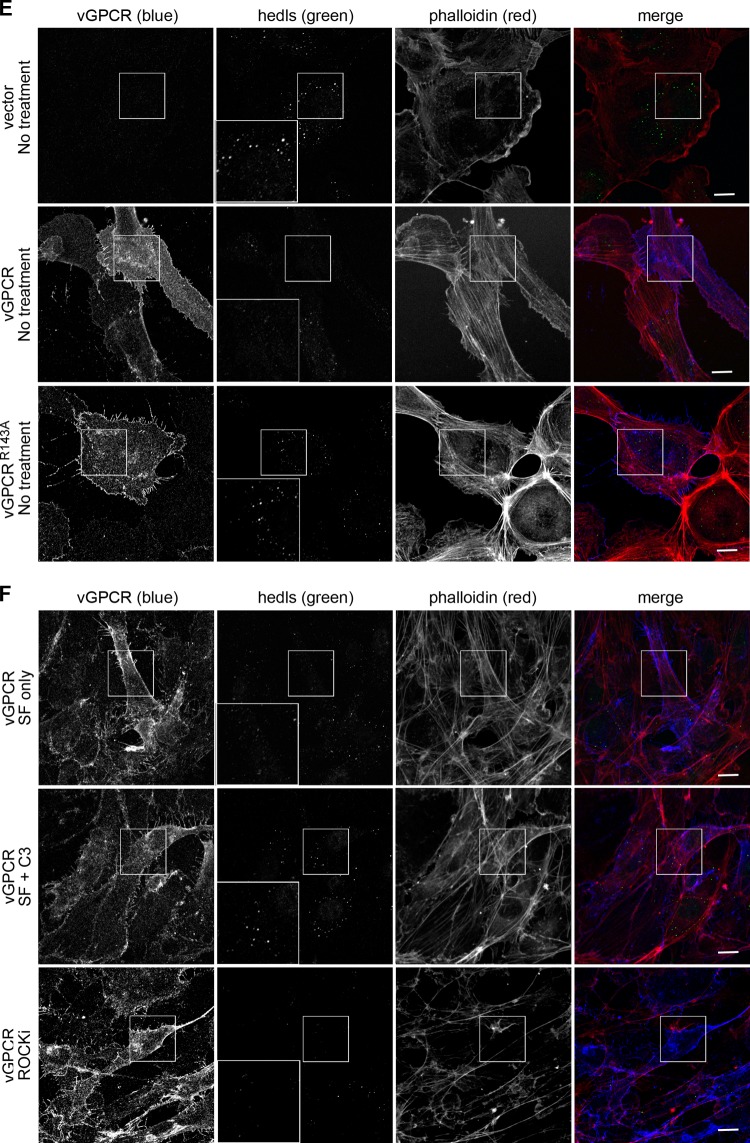
Fig 5.
vGPCR disrupts the formation of cytoplasmic PBs in a Rho-dependent fashion. (A) HUVE cells were treated with arsenite for 30 min to induce stress and the formation of PBs. After fixation and permeabilization, PBs were immunostained with two antibodies directed against resident proteins: anti-hedls antibody (green), which also binds to nuclear S6K, and anti-DDX6 antibody (red). (B and C) HUVE cells, transduced with empty vector and selected as described in Fig. 2, were treated with the RhoA activators lysophosphatidic acid (LPA) (B, C), nocodazole (C), or Rho activator II (C) as described in Materials and Methods. After fixation, cells were stained with anti-hedls antibody (green) to visualize PBs, and Alexa 555-conjugated phalloidin (red) to visualize actin. (C and D) To quantify the effect of Rho activation on PB accretion, the number of cells with normal (approximately 1 μm in diameter)-sized PBs per each field of view was counted. After transduction with vGPCR, the number of vGPCR-positive cells with normal PBs was counted. For each treatment, 100 to 200 cells were counted from three independent experiments. The fold reduction in the number of cells with PBs was calculated as described in Materials and Methods. (D to F) After transduction with vGPCR (wild type or signaling-inactive mutant R143A) or control retroviruses, selected cells were either mock treated in serum-free medium or treated with 1 μg/ml of C3 transferase, an irreversible inhibitor of RhoA subfamily GTPases, for 6 h, followed by 1 h in normal medium. Additional wells were treated with an inhibitor of the RhoA-associated kinase, ROCK, for 1 h, or not treated. These cells were fixed and triple stained with antibodies to the PB resident hedls protein (green) and vGPCR (falsely colored blue) and Alexa 647-conjugated phalloidin (red) to visualize actin stress fibers. Images are representative of two (R143A mutant) or five (vector and wild-type vGPCR) independent experiments (scale bar = 10 μm). Insets clearly indicate the presence or absence of PBs in vGPCR-expressing cells under different conditions.