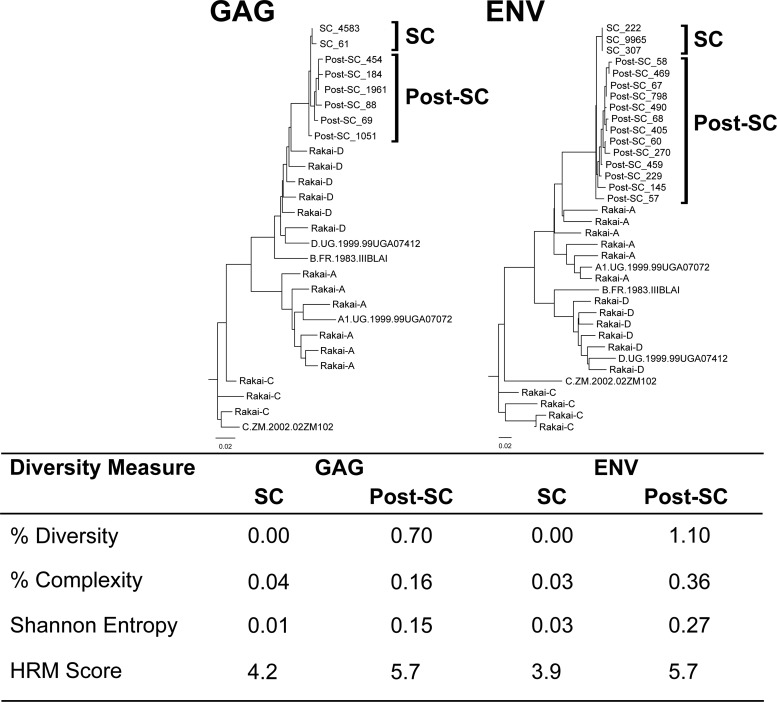
Fig 1.
Diversity data collected from a representative study participant. Neighbor-joining trees of HIV GAG and ENV next-generation consensus sequences from one individual at two time points are shown. The number of sequence reads included in each consensus sequence is shown at the end of the sequence, following an underscore. Subtype reference sequences and random sequences from unrelated individuals in Rakai are included. The diversity measures associated with both time points and regions are shown in the table below the trees. Note that this participant is infected with an A/D recombinant strain of HIV (A in env, D in gag).