Abstract
The genus Achromobacter currently is comprised of seven species, including Achromobacter xylosoxidans, an opportunistic and nosocomial pathogen that displays broad-spectrum antimicrobial resistance and is recognized as causing chronic respiratory tract infection in persons with cystic fibrosis (CF). To enable strain typing for global epidemiologic investigations, to clarify the taxonomy of “Achromobacter-like” strains, and to elucidate the population structure of this genus, we developed a genus-level multilocus sequence typing (MLST) scheme. We employed in silico analyses of whole-genome sequences of several phylogenetically related genera, including Bordetella, Burkholderia, Cupriavidus, Herminiimonas, Janthinobacterium, Methylibium, and Ralstonia, for selecting loci and designing PCR primers. Using this MLST scheme, we analyzed 107 genetically diverse Achromobacter isolates cultured from biologic specimens from CF and non-CF patients, 1 isolate recovered from sludge, and an additional 39 strains obtained from culture collections. Sequence data from these 147 strains, plus three recently genome-sequenced Achromobacter strains, were assigned to 129 sequence types based on seven loci. Calculation of the nucleotide divergence of concatenated locus sequences within and between MLST clusters confirmed the seven previously named Achromobacter species and revealed 14 additional genogroups. Indices of association showed significant linkage disequilibrium in all of the species/genogroups able to be tested, indicating that each group has a clonal population structure. No clear segregation of species/genogroups between CF and non-CF sources was found.
INTRODUCTION
When the genus Achromobacter was first proposed, it included Achromobacter xylosoxidans as its solitary species (17). Achromobacter ruhlandii and Achromobacter piechaudii subsequently were reassigned to this genus from the genus Alcaligenes (16), and thereafter, Achromobacter xylosoxidans subsp. denitrificans was reclassified as Achromobacter denitrificans (4). More recently, Achromobacter spanius, Achromobacter insolitus, and Achromobacter marplatensis were described as novel members of the genus (5, 7). These Achromobacter species vary with respect to their preferred environmental niches and impact on human health. A. xylosoxidans can be isolated from various water sources and is well recognized as an opportunistic human pathogen capable of causing a variety of infections, including endophthalmitis, keratoconjunctivitis, catheter-associated bloodstream infection, endocarditis, pneumonia, meningitis, and peritonitis (1, 12, 14). The remaining Achromobacter species are considered inhabitants of soil and have been much less frequently associated with human infection (4, 5, 7, 9).
Achromobacter infections are particularly noteworthy in persons with cystic fibrosis (CF). Although potential virulence factors remain to be elucidated and the role that respiratory tract infection with Achromobacter plays in contributing to lung disease in CF remains unclear, existing data indicate that airway infection in CF patients can be transient or chronic (10, 15). The incidence of Achromobacter infection in this patient population may be increasing; approximately 6.0% of CF patients were reported to have had Achromobacter infection in 2005, with the highest rate (8.1%) being seen among persons between the ages of 18 and 25 years (10).
It is a challenge to better understand the epidemiology, ecology, and clinical impact of human infection with Achromobacter because the taxonomy and population structure of these species are poorly defined, and consequently, accurate species identification is problematic. Achromobacter species are frequently misidentified by commercial identification systems, being most commonly mistaken for Alcaligenes, Bordetella, Burkholderia, Cupriavidus, Pandoraea, Ralstonia and Stenotrophomonas species (2, 11). By using repetitive-element PCR (rep-PCR)-based genotyping employing the BOX A1R primer (BOX-PCR), we have noted remarkable diversity among strains we previously identified as A. xylosoxidans using a species-specific PCR assay designed to target signature 16S rRNA sequences in this species (11).
The objective of this study was to develop a multilocus sequence typing (MLST) scheme for Achromobacter to elucidate the taxonomy and population structure of this genus. We took advantage of an extensive collection of diverse “Achromobacter-like” strains, as well as several recently available whole-genome sequences of strains belonging to related species.
MATERIALS AND METHODS
Bacterial strains.
The 150 strains included in this study are listed in the supplemental material (see Table S1). A total of 115 isolates recovered from culture of biologic specimens from patients with and without CF were obtained from the strain collection of the Burkholderia cepacia Research Laboratory and Repository (University of Michigan). All of these strains tested positive in a PCR assay previously designed to target Achromobacter 16S rRNA (11) and were selected, based on repetitive-element PCR genotyping (3), to represent distinct strains (i.e., to avoid including duplicate strains in the sample set). An additional strain was recovered from sludge. Thirty-one reference strains, which included representatives of the seven named Achromobacter species, were obtained from the BCCM/LMG (Ghent, Belgium) and ATCC (Manassas, VA) culture collections. DNA sequences from the three remaining Achromobacter strains were obtained from NCBI (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi).
Culture of bacteria and preparation of DNA.
All bacterial cultures were incubated aerobically at 32°C for 24 h on Mueller-Hinton agar. For DNA preparation, a single colony was suspended in 20 μl of lysis buffer containing 0.25% (vol/vol) sodium dodecyl sulfate and 0.05 N NaOH. After heating for 15 min at 95°C, 180 μl of high-pressure liquid chromatography (HPLC)-grade H2O was added. The suspension was centrifuged at 13,300 rpm for 5 min, and the supernatant was stored at 4°C.
Selection of MLST loci.
At the outset of this project, no Achromobacter whole-genome sequences were available. Thus, we initially analyzed annotated DNA sequences of phylogenetically related (based on 16S rRNA sequence analysis) genera available at NCBI, including the following 19 strains, representing seven genera: Bordetella avium 197N, Bordetella bronchiseptica RB50, Bordetella pertussis Tohama I, Bordetella parapertussis 12822, Bordetella petrii DSM 12804, Burkholderia ambifaria MC40-6, Burkholderia cenocepacia J2315, Burkholderia mallei ATCC 23344, Burkholderia phytofirmans PsJN, Cupriavidus metallidurans CH34, Cupriavidus necator H16, Cupriavidus pinotubonensis JMP134, Cupriavidus taiwanensis LMG 19424, Herminiimonas arsenicoxydans ULPAs1, Janthinobacterium sp. Marseille, Methylibium petroleiphilum PM1, Ralstonia pickettii 12D, Ralstonia pickettii 12J, and Ralstonia solanacearum GMI1000.
DNA sequences were aligned with MAUVE 2.3.0 build 98(c) using default parameters with MUSCLE 3.6 to identify large contiguous blocks (LCBs) that were present in all genomes and that had approximately 70% homology. LCBs were analyzed, using the annotated genome of B. bronchiseptica RB50 as a reference, to identify candidate MLST genes. Candidate genes from all 19 sequenced strains were aligned using Clustal W, and the resulting dendrograms were each compared to a dendrogram based on the respective 16S rRNA genes from these strains. If the two dendrograms indicated different phylogenies, then the gene was eliminated as a possible MLST candidate. Candidate genes were limited to those with only a single copy in the genome, and in the case of species with multiple chromosomes (e.g., Burkholderia spp.), candidate genes were also limited to those located on the primary chromosome. Genes were removed as candidates if a paralog or homolog with a nucleotide identity of ≥80% was identified in any one of the 19 sequenced strains. Ultimately, this process identified seven genes for inclusion in the MLST scheme: eno, gltB, lepA, nrdA, nuoL, nusA, and rpoB.
Amplification and DNA sequencing.
The MLST locus within each of the selected genes ranged in length from 214 bp (eno) to 449 bp (nrdA) (Table 1). PCR primers for each locus were manually designed to target sequences ∼50 to 200 bases upstream or downstream from the sequence of interest and then checked for hairpin turns and primer dimers using Primer Select (DNAStar, Madison, WI). Amplification of targeted DNA was carried out in 25-μl reaction volumes, each containing final concentrations of the following reagents: 2 mM MgCl2, 50 mM KCl, 20 mM Tris-Cl (pH 8.4), 250 μM (each) deoxynucleoside triphosphates (ISC Bio-Express, Kaysville, UT), 0.4 μM (each) primer (IDT Technology, Coralville, IA), 5% (vol/vol) dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO), 1 M betaine monohydrate (Sigma), 2 U of Taq polymerase (Invitrogen, Carlsbad, CA), and 2 μl of whole-cell bacterial lysate, adjusted to 25 μl by the addition of HPLC-grade H2O. Amplification was carried out in a PTC-100 (Bio-Rad, Hercules, CA) thermal controller. After an initial denaturation for 2 min at 95°C, 30 cycles were completed, with each consisting of 30 s at 94°C, 30 s at the appropriate annealing temperature (Table 1), and 60 s at 72°C. A final extension of 5 min at 72°C was applied with an infinite hold at 8°C. Amplified PCR products were purified using the Qiagen QIAquick PCR purification kit (Qiagen Inc., Valencia, CA) by following the manufacturer's instructions. DNA sequencing was carried out with an Applied Biosystems ABI model 3730 sequencer using the protocols provided by the manufacturer (PE Applied Biosystems, Foster City, CA) by using the BigDye Terminator cycle sequencing ready reaction kit.
Table 1.
Genes used for the MLST scheme with primers used for amplification
| Gene | Primer sets (5′ → 3′)a | Temp (°C) | Amplicon size (bp) | Gene fragment size (bp)b |
|---|---|---|---|---|
| nusA | P1, GCCTGCAAGARCCCGACAAGP2, GTCCATSGCGTGCTTGTCTTC | 59 | 795 | 355 |
| rpoB | P1, TGCCMTGGAACGGYTACAACP2, GGCCAGRTASACCTTGATCATCTT | 57 | 791 | 413 |
| eno | P1, ATGCCCGTGCCSATGATGAAP2, TCAGGGTGCCGATCTGGTTG | 57 | 613 | 214 |
| gltB | P1, TGCAACCGGGCAAGATGTTP2, TCGGACACGATCAGGATGTT | 57 | 685 | 241 |
| lepA | P1, CTAYAACCTGAACCTGATCGACACP2, GCGACTTSGGCGTGAACAC | 57 | 524 | 347 |
| nuoL | P1, CATGCACCAYRACCAGGACATP2, CGCGAACGCGTAGTGATAGATG | 56 | 880 | 230 |
| nrdA | P1, GAACTGGATTCCCGACCTGTTCP2, TTCGATTTGACGTACAAGTTCTGG | 56 | 954 | 449 |
Standard mixed oligonucleotide bases for primer sequences are listed in bold.
Sequence length used in MLST analysis for each locus.
Sequence chromatograms were visualized and edited with Chromas version 2.31 (Technelysium Pty. Ltd.). All sequences were aligned and appropriate cutoffs were applied using MegAlign (DNAStar); trimmed sequences were saved individually. Concatenation of sequences was performed with SeqEdit (DNAStar).
Allele and ST assignment.
Alignments of sequences from each gene were performed using Clustal V. A different number was assigned to each unique allele, and a sequence type (ST) was assigned to each unique allelic profile. These are available in the Achromobacter MLST database (http://pubmlst.org/achromobacter/).
Phylogenetic analyses.
Alignment of concatenated MLST sequences (2,249 bp) for each isolate was performed using MegAlign with Clustal W to generate a phylogenetic tree, with 1,000 bootstrap replications using the default parameters in MegAlign. Statistical analyses of allelic profiles, number of polymorphic sites, frequencies of alleles, GC content, and the ratio of nonsynonymous to synonymous substitutions (dN/dS) were calculated using START (http://pubmlst.org/software/analysis/start/). The standardized index of association (IA) was calculated using LIAN 3.5 (http://pubmlst.org), and significance (P = 0.05) was established under the null hypothesis of linkage equilibrium (H0:VD = Ve) by performing 1,000 Monte Carlo resamplings. Simpson's index of diversity was calculated using the comparing partitions program (available at http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Tool).
For validation of distinctness of sequence similarity clusters in the population, the k parameter, which is the ratio of the intergroup divergence to the mean of the intragroup divergence, was calculated as previously described (13). Ratios greater than 2 were considered distinct sequence similarity clusters. Subgroups within clusters were analyzed separately and designated “a” or “b.”
All sequence data and related information were deposited in the Achromobacter MLST database (http://pubmlst.org/achromobacter/).
RESULTS
Allelic variation in Achromobacter.
Analysis of all 150 strains revealed 129 STs (see Table S1 in the supplemental material). The number of alleles at each locus ranged from 53 (gltB) to 70 (rpoB), while the number of polymorphic sites ranged from 45 (gltB) to 93 (lepA) (Table 2). Simpson's index of diversity ranged from 0.959 (nuoL) to 0.980 (nusA), indicating a high level of discrimination. Low dN/dS values indicated the absence of strong positive selection pressure at these loci and supported their suitability for use in population genetic analyses.
Table 2.
Analysis of the seven MLST loci in the 150 strains studied
| Gene | No. of alleles | No. of polymorphic sites | % GC content | Simpson's index (D) | dN/dS |
|---|---|---|---|---|---|
| nusA | 62 | 87 | 64.6 | 0.973 | 0.0136 |
| rpoB | 70 | 52 | 62.8 | 0.959 | 0.0448 |
| eno | 64 | 47 | 60.6 | 0.973 | 0.0469 |
| gltB | 53 | 45 | 62.9 | 0.963 | 0.0350 |
| lepA | 69 | 93 | 64.9 | 0.978 | 0.0946 |
| nuoL | 58 | 54 | 62.2 | 0.976 | 0.1042 |
| nrdA | 65 | 77 | 58.2 | 0.980 | 0.0473 |
Achromobacter species and genogroup delineations.
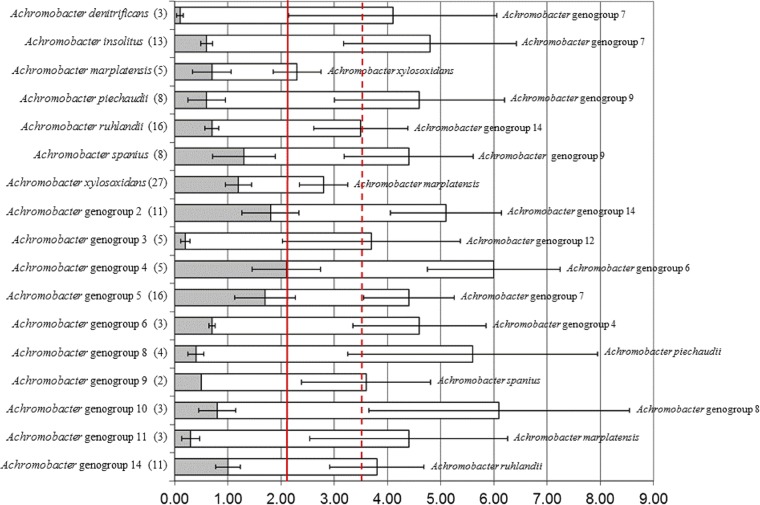
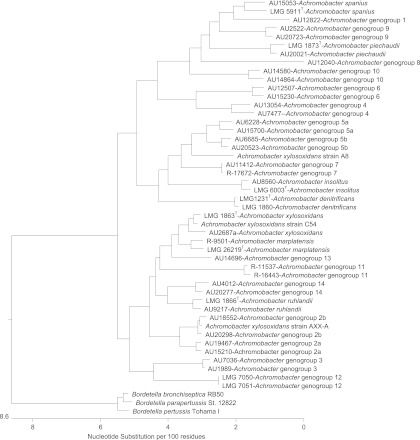
Alignment of concatenated MLST sequences from all 150 isolates revealed interspecies/genogroup divergence rates ranging from 1.6% to 5.3%. Therefore, a rate of 1.6% was used as the cutoff for defining the lower limit of any single species or genogroup (Fig. 1). Maximum intraspecies divergence was 2.1%. Applying this as the cutoff in the concatenated MLST dendrogram (rooted with Bordetella reference strains) identified at least 14 additional genogroups, most of which had distinctness of population ratio (k) parameters of ≥2 (Fig. 2). A. xylosoxidans and A. marplatensis could not be considered separate sequence similarity clusters, with k values of 1.68, despite both being taxonomically validly named species. The only other genogroup failing to meet the distinctness of population ratio was Achromobacter genogroup 5 (k = 1.58); however, the closest genogroup, Achromobacter genogroup 7, contained only 2 strains. The five remaining previously named Achromobacter species and new genogroups had k parameters indicating distinctness (range, 2.36 to 40.00).
Fig 1.
Nucleotide divergence of concatenated MLST loci. Shaded bars represent divergence within a species/genogroup; open bars represent the divergence between species/genogroup and its closest neighbor (listed to the right), as defined by percentage similarity. Error bars represent the standard errors of the shaded and open bars. The solid line represents maximum intraspecies divergence; the dashed line represents the average interspecies divergence.
Fig 2.
Dendrogram of concatenated MLST loci sequences containing two strains of each Achromobacter species or genogroup (unless only a single strain was available) and rooted with Bordetella species.
Presence of distinct subgroups.
The concatenated MLST tree revealed that Achromobacter genogroups 2 and 5 contained distinct subgroups, each labeled a and b. Using bootstrapping with 1,000 replicates, Achromobacter genogroups 2a and 2b showed 100% support that distinct nodes would be maintained. Similarly, Achromobacter genogroups 5a and 5b showed 93.8% support that distinct nodes would be maintained. Intradivergence and interdivergence levels, as well as k parameters, indicated that genogroups 2a and 2b and genogroups 5a and 5b could be considered separate clusters, with k parameters of 3.27 and 2.62, respectively. These data provide evidence that these two genogroups contain distinct diverging subgroups.
Linkage disequilibrium.
Assessment of the population structure of Achromobacter was investigated by calculating the IA for each species or genogroup. All species/genogroups showed IA values differing from zero, indicating that all were in linkage disequilibrium. However, five groups (genogroups 1, 7, 9, 12, and 13) did not include enough strains for reliable testing of significance between the observed variance and maximum trial variance based on the criteria proposed by Kaiser et al. (8). Among the remaining 16 species/genogroups, all showed significant evidence of linkage disequilibrium based on the number of strains analyzed, while 14 maintained significance when analyzed for number of STs only (see Table S2 in the supplemental material).
DISCUSSION
Achromobacter species are increasingly recognized as nosocomial and opportunistic human pathogens, particularly in persons with CF, in whom respiratory tract infection likely contributes to progressive lung disease (10). The broad-spectrum antibiotic resistance of most clinical isolates limits effective therapy of infection, and prevention of infection and identification of virulence factors are challenged by an incomplete understanding of the ecology, epidemiology, and taxonomy of Achromobacter species. Unfortunately, little progress has been made in these areas during the past decade. The novel species A. insolitus and A. spanius were added to the genus in 2003, and A. marplatensis was proposed as yet another novel species in 2010, bringing the total number of validly named species in this genus to seven (7). However, our ongoing analyses of bacterial isolates recovered primarily from human specimens have identified a variety of “Achromobacter-like” strains that cannot be placed into one of these seven species with confidence, suggesting the existence of several additional distinct taxa. These strains, along with strains from each of the seven named species, tested positive in a previously described PCR assay targeting 16S rRNA gene sequences intended to be specific for A. xylosoxidans (11). Genotyping analyses of such strains using a rep-PCR typing method (3) indicated a high level of genetic diversity and identified several clusters, again suggestive of distinct species.
In order to delineate the taxonomy and epidemiology of Achromobacter species, we sought to devise an MLST scheme that would not only provide strain genotyping but also enable reliable genus-level phylogenetic analyses. We took advantage of the whole-genome sequences available for 19 strains from several related genera and employed a strategy to select candidate MLST genes that would predict phylogenetic relationships among these genera concordant with those predicted by analysis of their complete 16S rRNA gene sequences. Limiting MLST genes to those located on the primary chromosome in species with multiple chromosomes (e.g., Burkholderia spp.) avoided genes with potentially greater rates of evolutionary change that may be found on secondary chromosomes (6).
We applied this MLST scheme to examine a large collection of putative Achromobacter strains, primarily recovered from human sources and selected based on previous genotyping analysis to represent a diverse strain set. The analysis reiterated the high level of diversity in this set, reflected by a large number of alleles and polymorphic sites, as well as a Simpson index of diversity of at least 0.959 in all seven loci. Our results revealed the existence of 14 distinct genogroups in addition to the seven currently named species. Analysis of the concatenated MLST sequences from the 150 strains included in the study demonstrated that these 21 species/genogroups had low (maximum, 2.1%) intragroup and high (average, 3.54%) intergroup sequence divergence levels. The k parameter, a measure of distinctness between two groups, was high (>2) for most pairwise sets; the value between genogroup 5 and genogroup 7 was limited by the inclusion of only two strains in genogroup 7. Interpretation of the low k value (1.68%) between A. xylosoxidans and A. marplatensis was similarly limited by the presence of only two distinct strains in the latter species. This species has been shown, nevertheless, to be a distinct Achromobacter species on the basis of polyphasic analyses, including DNA-DNA hybridization values and phenotypic characteristics (7). The concatenated MLST analysis also identified two distinct subgroups in both genogroup 2 and genogroup 5. Sequence divergence levels, as well as k values, indicated that these four subgroups most likely represent distinct taxa. Confirmation of this will require further taxonomic analyses and would corroborate that the design of our MLST scheme provides phylogenetically sound data.
The IA, a summary statistic based on the variance of pairwise distances between strains in a set, revealed values different from zero for all 21 species/genogroups. However, because the expected value of IA depends on sample size, testing of the significance of the IA was not reliable in the five genogroups that each contained fewer than three strains. We found significant evidence for linkage disequilibrium in all remaining groups, including the subgroups in genogroups 2 and 5, consistent with a clonal epidemic population structure.
Our results have important implications in efforts to better understand the epidemiology, ecology, clinical microbiology, and virulence of Achromobacter species. Advances in each of these areas are predicated on a clearer appreciation of the taxonomy and population structure of this genus. For example, we have found that, based on a preliminary analyses of a large set of Achromobacter isolates recovered from respiratory cultures from CF patients in the United States, approximately 1/3 are not A. xylosoxidans, although essentially all were initially identified as this species by commercial identification systems. It appears A. ruhlandii will comprise a considerable minority of Achromobacter species recovered from CF patients. Our analysis also shows that, among the A. xylosoxidans strains for which genome sequencing has been performed, only strain C54 is actually a member of this species; strain AXX-A is placed in genogroup 2b, while strain A8 cannot be confidently placed in any of the 21 species/genogroups identified in our study.
Our MLST scheme provides a sequence database and method for genotyping Achromobacter strains for global epidemiologic studies and for assigning strains to defined or new genogroups. Ongoing work is aimed at developing less labor-intensive methods to delineate each of the species/genogroups in this genus.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Cystic Fibrosis Foundation.
Footnotes
Published ahead of print 11 July 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aisenberg G, Rolston KV, Safdar A. 2004. Bacteremia caused by Achromobacter and Alcaligenes species in 46 patients with cancer (1989–2003). Cancer 101:2134–2140 [DOI] [PubMed] [Google Scholar]
- 2. Brisse S, et al. 2002. Comparative evaluation of the BD Phoenix and VITEK 2 automated instruments for identification of isolates of the Burkholderia cepacia complex. J. Clin. Microbiol. 40:1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coenye T, Spilker T, Martin A, LiPuma JJ. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coenye T, et al. 2003. Kerstersia gyiorum gen. nov., sp. nov., a novel Alcaligenes faecalis-like organism isolated from human clinical samples, and reclassification of Alcaligenes denitrificans Ruger and Tan 1983 as Achromobacter denitrificans comb. nov. Int. J. Syst. Evol. Microbiol. 53:1825–1831 [DOI] [PubMed] [Google Scholar]
- 5. Coenye T, Vancanneyt M, Falsen E, Swings J, Vandamme P. 2003. Achromobacter insolitus sp. nov. and Achromobacter spanius sp. nov., from human clinical samples. Int. J. Syst. Evol. Microbiol. 53:1819–1824 [DOI] [PubMed] [Google Scholar]
- 6. Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. 2010. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput. Biol. 6:e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gomila M, et al. 2011. Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol-contaminated soil. Int. J. Syst. Evol. Microbiol. 61:2231–2237 [DOI] [PubMed] [Google Scholar]
- 8. Kaiser S, Biehler K, Jonas D. 2009. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 191:2934–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kiredjian M, Holmes B, Kersters K, Guilvout I, De Ley J. 1986. Alcaligenes piechaudii, a new species from human clinical specimens and the environment. Int. J. Syst. Evol. Microbiol. 36:282–287 [Google Scholar]
- 10. LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu L, et al. 2002. Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 40:1210–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oh JY, Shin YJ, Wee WR. 2005. A case of epidemic keratoconjunctivitis complicated by Alcaligenes xylosoxidans infection. Korean J. Ophthalmol. 19:233–234 [DOI] [PubMed] [Google Scholar]
- 13. Palys T, Nakamura LK, Cohan FM. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145–1156 [DOI] [PubMed] [Google Scholar]
- 14. Robert PY, et al. 2008. Alcaligenes xylosoxidans endophthalmitis following phacoemulsification and intraocular lens implantation. Ophthalmic Surg. Lasers Imaging 39:500–504 [DOI] [PubMed] [Google Scholar]
- 15. Rønne Hansen C, Pressler T, Høiby N, Gormsen M. 2006. Chronic infection with Achromobacter xylosoxidans in cystic fibrosis patients; a retrospective case control study. J. Cyst. Fibros. 5:245–251 [DOI] [PubMed] [Google Scholar]
- 16. Yabuuchi E, Kawamura Y, Kosako Y, Ezaki T. 1998. Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Ruger and Tan) comb. nov. Microbiol. Immunol. 42:429–438 [DOI] [PubMed] [Google Scholar]
- 17. Yabuuchi E, Yano I. 1981. Achromobacter gen. nov. and Achromobacter xylosoxidans (ex Yabuuchi and Ohyama 1971) nom. rev. Int. J. Syst. Evol. Microbiol. 31:477–478 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.