Abstract
Infection with human cytomegalovirus (HCMV) continues to be a major threat for pregnant women and the immunocompromised population. Although several anti-HCMV therapies are available, the development of new anti-HCMV agents is highly desired. There is growing interest in identifying compounds that might inhibit HCMV by modulating the cellular milieu. Interest in cardiac glycosides (CG), used in patients with congestive heart failure, has increased because of their established anticancer and their suggested antiviral activities. We report that the several CG—digoxin, digitoxin, and ouabain—are potent inhibitors of HCMV at nM concentrations. HCMV inhibition occurred prior to DNA replication, but following binding to its cellular receptors. The levels of immediate early, early, and late viral proteins and cellular NF-κB were significantly reduced in CG-treated cells. The activity of CG in infected cells correlated with the expression of the potassium channel gene, hERG. CMV infection upregulated hERG, whereas CG significantly downregulated its expression. Infection with mouse CMV upregulated mouse ERG (mERG), but treatment with CG did not inhibit virus replication or mERG transcription. These findings suggest that CG may inhibit HCMV by modulating human cellular targets associated with hERG and that these compounds should be studied for their antiviral activities.
INTRODUCTION
Human cytomegalovirus (HCMV) is a major pathogen in transplant recipients, HIV-infected individuals and congenitally infected children (6, 11, 24). HCMV replication has been associated with the outcome of several syndromes in immunocompetent individuals, including sepsis/pulmonary complications in patients in intensive care units and the brain tumor glioblastoma multiforme (28, 32, 34). With broadening indications for HCMV therapy, the side effects associated with anti-HCMV compounds (all viral DNA polymerase inhibitors), and the emergence of resistant viral mutants during therapy (5, 19, 46), there is a need to develop anti-HCMV compounds with novel mechanisms of action. Strategies for development of anti-HCMV agents include the identification of both viral and cellular targets that can abrogate virus replication (“anticellular antiviral” approach) (8, 10, 51).
Cardiac glycosides (CG) have been reported to inhibit herpes simplex virus 1 (HSV-1) and HCMV (7, 13). These compounds are prescribed for congestive heart failure, a condition in which they bind to and inhibit the activity of the Na+,K+-ATPase pump (31). Effects other than inhibition of the Na+,K+-ATPase are becoming evident, mainly anticancer activities (31, 47, 52). Inhibition of cancer cell growth likely results from modulation of cell signaling pathways and possibly from interaction with other ion channels. A subgroup of voltage-gated K+ channels, encoded by the hERG gene, which is usually not expressed in normal cells, is upregulated in cancer cells (2, 35, 36). CG were reported to inhibit hERG trafficking, not via direct interaction with hERG, but possibly through secondary interaction with protein kinases/chaperones that are important in hERG processing (9, 50). The role of hERG in HCMV replication has not been studied. We investigated the characteristics of HCMV inhibition by two CG, digoxin and ouabain, and the potential role of hERG in HCMV inhibition by CG.
MATERIALS AND METHODS
Compounds.
Digoxin, ouabain, digitoxin, and ganciclovir (GCV) were purchased from Sigma Chemicals (St. Louis, MO). The CG were dissolved in dimethyl sulfoxide (DMSO), and GCV was dissolved in distilled water. Stock solutions (10 mM) were stored at −80°C.
Viruses.
The pp28-luciferase HCMV Towne strain was constructed as previously described (15). This virus expresses luciferase under the control of the pp28 late promoter. Luciferase expression is strongly activated at 48 to 72 h postinfection (hpi). This reporter system is sensitive and reproducible and closely correlates with plaque reduction (15). We generated a pp28-luciferase GCV-resistant strain; the C607Y mutation in UL97 was confirmed by sequence analysis (30). A luciferase-tagged HSV-1 strain (KOS/Dlux/oriS) was used to evaluate the inhibition of HSV-1. Mouse CMV (MCMV) Smith strain and Epstein-Barr virus-positive (EBV+) Akata cells were used to evaluate the inhibition of MCMV and EBV, respectively.
Cell culture, virus infection, and antiviral assays.
Human foreskin fibroblasts (HFFs), passages 12 to 16 (ATCC, CRL-2088), and U373 glioma cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA) in a 5% CO2 incubator at 37°C and used for infection with HCMV at a multiplicity of infection (MOI) of 1 PFU/cell. After 90 min of adsorption, the medium was removed, and the cells were washed with phosphate-buffered saline (PBS). DMEM with 4% FBS containing compounds was added to each well. Infected, treated HFFs were collected at 72 hpi, and lysates were assayed for luciferase using a luciferase assay kit (Promega, Madison, WI) on a GloMax-Multi+ detection system (Promega) according to the manufacturer's instructions. Infection with HSV-1–luciferase was performed according to the principles described for HCMV, except that luciferase was quantified at 24 hpi. MCMV was diluted in DMEM to a concentration that gave ∼100 plaques per well. After the infection of mouse embryonic fibroblasts (MEFs), compounds were added, and a methylcellulose overlay was applied to each well. After 3 days of incubation, the cells were stained with crystal violet, and plaques were counted under a microscope at ×40 magnification. Akata cells latently infected with EBV were cultured in RPMI 1640 medium (Sigma) supplemented with 10% FBS. The cells were induced with 50 μg/ml of goat antihuman IgG (Sigma). Compounds were added to triplicate wells immediately after lytic induction. Supernatants were collected after 4 days for DNA quantification by real-time PCR (27).
Transient transfection.
For overexpression of hERG, U373 cells were transfected with hERG-pcDNA3.1 plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The pcDNA3.1 plasmid is derived from pcDNA3 and provides high-level transient expression in mammalian cells. The vector contains an HCMV immediate-early (IE) promoter for high-level expression. Cells were seeded onto six-well tissue culture plates. After incubation at 37°C overnight, the cells were transfected with hERG-pcDNA3.1 (1 μg/well) for 6 h; the medium was then replaced, and the cells were infected with HCMV and treated with compounds. After 24 h of incubation, the cells were lysed for Western blotting.
To investigate the effect of CG on CMV promoter activity, an enhanced green fluorescent protein (EGFP) reporter plasmid, pEGFP-C (Clontech, Mountain View, CA), was used. This plasmid expresses EGFP under the control of a CMV IE promoter, the same promoter that controls the expression of hERG cDNA in pcDNA3.1 plasmid. U373 cells were transiently transfected with pEGFP-C (1 μg/well) using Lipofectamine 2000. After 6 h, the medium was replaced, and the cells were infected with HCMV and treated with compounds. EGFP expression was determined by Western blotting at 24 hpi. To further investigate the effect of CG on CMV promoter activity, pRL Renilla luciferase control reporter plasmid (Promega) was used. The pRL-CMV vector (accession no. AF025843) contains CMV enhancer and IE promoter elements. For transient transfection, U373 cells were plated onto 24-well plates and transfected with pRL-CMV plasmid. After 6 h, the cells were treated with the compounds. The Renilla luciferase activity was measured after 24 h by mixing cell lysates with a Renilla luciferase substrate, coelentrazine (Sigma), dissolved in PBS.
Cell viability.
Cell viability was determined by an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide]-based colorimetric assay. Cells were seeded in 96-well microplates and treated with various concentrations of CG. For each cell type used for virus infection and drug treatment, an MTT assay was performed at the same time points of the antiviral assay.
SDS-PAGE and immunoblot analysis.
Cell lysates containing equivalent amount of proteins were mixed with an equal volume of sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 5% β-mercaptoethanol) and boiled at 100°C for 10 min. Denatured proteins were resolved in Tris-glycine polyacrylamide gels (10 to 12%) and transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA) by electroblotting. Membranes were incubated in blocking solution (5% nonfat dry milk and 0.1% Tween 20 in PBS [PBST]) for 1 h, washed three times with PBST, and incubated with appropriately diluted primary antibodies at 4°C overnight. The membranes were washed with PBST, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies in PBST for 1 h at room temperature. After a washing step with PBST, protein bands were visualized by chemiluminescence using SuperSignal West Dura and Pico reagents (Pierce Chemical, Rockford, IL). The following antibodies were used: mouse anti-IE1 and -IE2 antibody (MAb810), mouse anti-β-actin antibody (Millipore, Billerica, MA), mouse anti-UL84 antibody, mouse anti-NF-κB antibody (p65), mouse anti-UL44 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-pp65 antibody (Vector Laboratories, Burlingame, CA), HRP-conjugated anti-rabbit IgG antibody (Cell Signaling Technology, Beverly, MA), rabbit anti-KV11.1 (hERG) antibody (Alomone Labs, Jerusalem, Israel), rabbit anti-GFP antibody (G1544; Sigma), and HRP-conjugated anti-mouse IgG (GE Healthcare, Waukesha, WI). Individual bands intensity was quantified by densitometry using ImageJ v1.41o software (National Institutes of Health, Bethesda, MD).
RNA isolation and RT-PCR.
Total RNA was isolated from cultured cells by using an RNeasy minikit (Qiagen, Georgetown, MD). A RevertAid first-strand cDNA synthesis kit (Fermentas Life Sciences, Cromwell Park, MD) was used to synthesize first-strand cDNA from total RNA using oligo(dT) primers. Synthesis of first-strand cDNA from mRNA template was carried out at 42°C for 1 h. The levels of hERG1 and hERG1B isoforms were detected by using sequence-specific primers and PCR (94°C for 30 s, 55°C for 30 s, and 72°C for 60 s for 30 cycles). PCR products were analyzed on 2% agarose gel and stained with ethidium bromide (Sigma). In addition, quantitative reverse transcription-PCR (qRT-PCR) was performed using specific primers and SYBR green (Fermentas Life Sciences) with a two-step cycling protocol (95°C for 15 s and 60°C for 1 min). The transcription of three other potassium channel genes (Kir1.1, Kir 2.1, and KCNQ5) and mERG was quantified. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control. Primer sequences for qualitative and quantitative PCR may be found in the supplemental material.
HCMV real-time PCR.
The inhibitory effects of CG on HCMV DNA synthesis were determined by real-time PCR. DNA was isolated from infected and treated cells at 48, 72, and 96 hpi. The real-time PCR is based on detection of the highly conserved US17 gene (49).
Drug combination.
The combined inhibitory effect of CG and GCV on HCMV replication was determined in infected HFFs. Compounds were added at different dilutions, starting with concentrations that resulted in full inhibition. Analysis was performed using the Bliss equation in which drug combination represents the product of two probabilistically independent events (21):
where D is the drug concentration, m is the slope, and EC50 is the effective concentration resulting in 50% virus inhibition. The combined effect of two inhibitors (FU, fractional unaffected) is computed as the product of individual effects of the two inhibitors, FU1 and FU2. If the ratio of observed fold inhibition divided by the expected fold inhibition is greater than 1, the compounds are synergistic. If the ratio is less than 1, the combination is considered antagonistic, and if it is equals to 1 the combination is additive.
RESULTS
CG are potent inhibitors of HCMV.
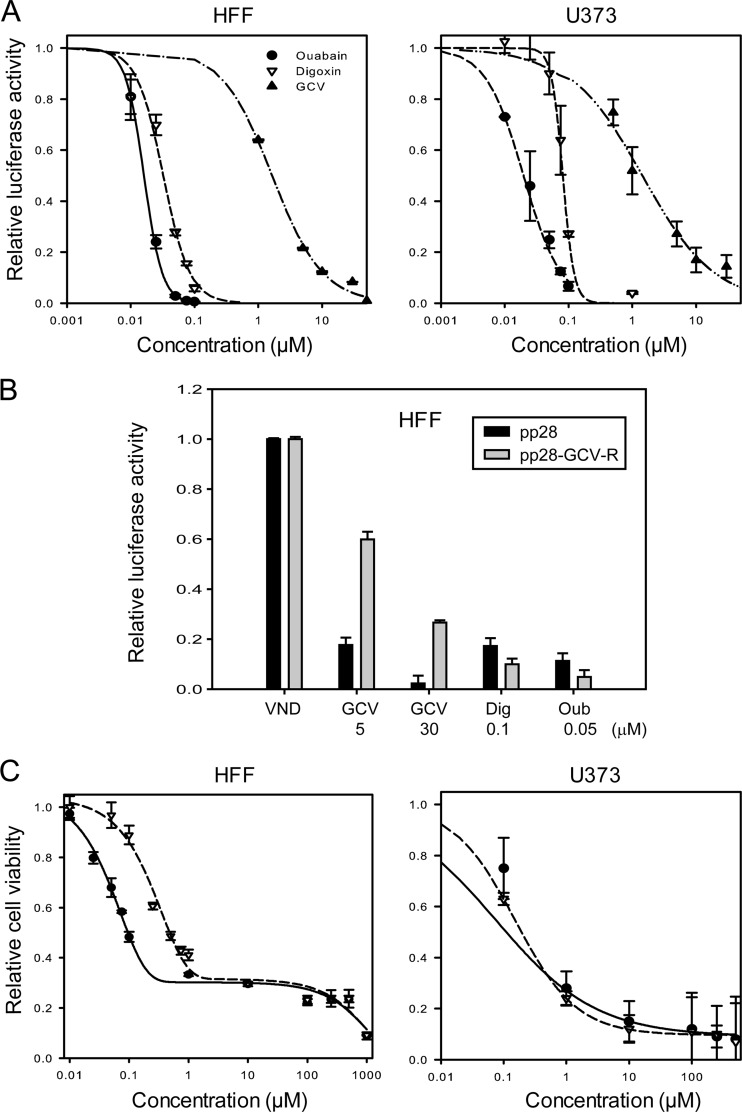
HFFs and U373 cells were infected with Towne HCMV luciferase and treated with digoxin and ouabain at 0.01 to 0.1 μM. GCV was used as a control (1 to 30 μM). Virus replication was quantified by luciferase activity at 72 hpi. Treatment with digoxin and ouabain resulted in a significant inhibition of HCMV replication in HFFs and U373 cells (Fig. 1A and Table 1). In HFFs, the 50% effective concentrations (EC50s) of digoxin and ouabain were 0.036 μM ± 0.00 and 0.017 μM ± 0.00, respectively. The EC50 of GCV was 1.4 μM ± 0.01, and the slope of its dose-response curve was 1, as expected from a compound with one target. The slope of CG was >2, suggesting an involvement of one target with subsequent downstream effects or multiple targets (44). The GCV-resistant Towne HCMV was inhibited by CG, while GCV showed decreased activity against the resistant virus (Fig. 1B).
Fig 1.
CG inhibit HCMV replication. (A) HFF and U373 cells were infected with pp28-luc HCMV and treated with the indicated concentrations of digoxin, ouabain, or GCV. The luciferase activity was measured at 72 hpi. The data represent mean values ± the SD of triplicate determinations from five independent experiments. (B) HFF cells were infected with either pp28-luc HCMV or a GCV-resistant pp28-luc HCMV (pp28-GCV-R) and treated with the indicated concentrations of digoxin, ouabain, or GCV. The luciferase activity was determined at 72 hpi. VND, virus/no drug. The data represent mean values ± the SD of triplicate determinations from two independent experiments. (C) HFFs and U373 cells were treated with the indicated concentrations of digoxin and ouabain, and cell viability was determined after 72 h using an MTT assay. The data represent mean values ± the SD of triplicate determinations from three independent experiments.
Table 1.
Inhibition of alpha herpesvirus (HSV-1), betaherpesvirus (HCMV), and gammaherpesvirus (EBV) by CGsa
| Virus | Mean EC50 (μM) ± SD |
Mean CC50 (μM) ± SD |
SI |
Host cell | |||
|---|---|---|---|---|---|---|---|
| Digoxin | Ouabain | Digoxin | Ouabain | Digoxin | Ouabain | ||
| HCMV | 0.036 ± 0.00 | 0.017 ± 0.00 | 0.45 ± 0.11 | 0.10 ± 0.02 | 12.6 | 6.08 | HFF |
| HCMV | 0.086 ± 0.01 | 0.021 ± 0.00 | 0.48 ± 0.24 | 0.21 ± 0.07 | 5.6 | 10 | U373 |
| HSV-1 | 0.10 ± 0.01 | 0.026 ± 0.00 | 0.89 ± 0.10 | 0.58 ± 0.01 | 8.9 | 22 | Vero |
| EBV | 0.15 ± 0.00 | 0.077 ± 0.01 | 0.217 ± 0.05 | 0.10 ± 0.00 | 1.45 | 1.31 | Akata |
The EC50, CC50, and selectivity index (SI) values of digoxin and ouabain against HCMV, HSV-1, and EBV in different cell lines are shown. The EC50 of digoxin against HCMV in HFF cells has been reported to be 0.02 μM (13). The EC90s of digoxin and ouabain against HSV-1 in Vero cells were 0.15 and 0.04 μM, respectively (7). Mean values of triplicate determinations from three independent experiments are shown.
In HFFs the viability reached a plateau at 1 μM digoxin and 0.5 μM ouabain and remained unchanged despite increasing drug concentrations to 500 μM. At 1 mM, both digoxin and ouabain showed nearly 90% cytotoxicity (Fig. 1C). In U373 cells the viability decreased as the doses were increased. We calculated the selectivity indices (SIs) of digoxin and ouabain based on the effective drug concentration that results in 50% virus inhibition (EC50) and the drug concentration that leads to 50% cellular cytotoxicity (CC50) (Table 1). The SI was determined as the ratio between CC50 and EC50. The reported values of EC50 and CC50 represent the means and standard deviations (SD) of data derived from at least three independent experiments performed in triplicates. The curve-fitting toolbox, Matlab software (v7.10; Mathworks, Natick, MA), was used to determine EC50 and CC50 values using a four-parameter logistic regression.
Given the potent anti-HCMV activity of digoxin and ouabain, we tested digitoxin, another CG that has been studied as a potential anticancer agent (29). HCMV replication was significantly inhibited: the mean EC50 of digitoxin in HFFs was 0.016 μM ± 0.00, suggesting that CG are a class of HCMV inhibitors and that there might be differences in HCMV inhibition similar to their anticancer activities (25). The following experiments used digoxin and ouabain as a proof of concept for antiviral activities.
Inhibition of HSV-1 and EBV replication by CG.
HFFs were infected with luciferase HSV-1–KOS/Dlux/oriS (48) at an MOI of 1 and treated with digoxin and ouabain (0.01 to 0.5 μM). Virus replication was quantified by luciferase activity at 24 hpi. Lytic EBV replication was induced in Akata cells, and supernatants of treated cells (digoxin, 0.05 to 0.5 μM; ouabain, 0.025 to 0.5 μM) were collected after 4 days for real-time PCR of the EBV DNA polymerase gene, BALF5 (27). CG inhibited HSV-1 and possibly EBV replication. The most significant inhibition was observed for HCMV (Table 1), but because of the toxicity profile in the tested cell lines, the SI was not significantly different between HCMV and HSV-1. The toxicity of CG in Akata cells, along with reduced activity, resulted in a lower SI for EBV. Subsequent experiments focused on CMV (human and mouse) inhibition by CG.
Pretreatment with CG is not required for HCMV inhibition.
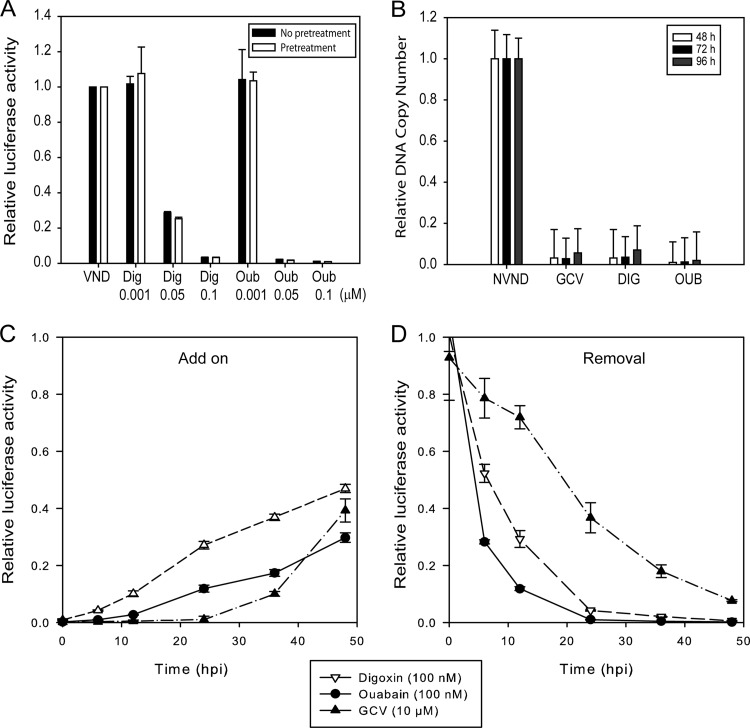
HFFs were incubated with CG 1 h prior to virus infection or after virus adsorption. The luciferase activity was measured at 72 hpi. Pretreated and nonpretreated cells showed similar degrees of virus inhibition (Fig. 2A), suggesting that CG were not acting at the stage of virus attachment or binding to cellular receptors.
Fig 2.
Timing of HCMV inhibition by CG. (A) Pretreatment with CG is not required for HCMV inhibition. HFFs were incubated with the indicated concentrations of digoxin or ouabain 1 h prior to virus infection in the presence of compounds (pretreatment). Another set of HFFs were infected and treated with the indicated concentrations of digoxin or ouabain (no pretreatment). The luciferase activity was measured at 72 hpi as an indication for late HCMV gene expression. VND, virus/no drug. The data represent mean values ± the SD of triplicate determinations from two independent experiments. (B) Viral DNA replication is inhibited by CG. HFFs were infected and treated with digoxin (0.1 μM), ouabain (0.05 μM), or GCV (10 μM), and viral DNA replication was quantified by real-time PCR at 48, 72, and 96 hpi. NVND, no virus/no drug. The data represent mean values the ± SD of triplicate determinations from two independent experiments. (C and D) CG inhibit HCMV during the first 24 h of virus replication. (C) HFFs were infected with pp28-luc HCMV, and compounds were added at 0, 6, 12, 24, 36, and 48 hpi (Add on). The luciferase activity was measured at 72 hpi. (D) HFF cells were infected with pp28-luc HCMV and treated with compounds immediately after virus adsorption. Compounds were removed at 0, 6, 12, 24, 36, and 48 hpi (Removal). The luciferase activity was measured at 72 hpi. The data represent mean values ± the SD of triplicate determinations from two independent experiments.
Inhibition of HCMV DNA replication by CG.
HFFs were infected and treated with digoxin (0.1 μM), ouabain (0.05 μM), or GCV (10 μM). Genomic DNA was purified from infected, treated cells after 48, 72, and 96 h, and the viral DNA replication was quantified by real-time PCR (49). Treatment with CG resulted in near-complete inhibition of HCMV DNA replication (Fig. 2B).
CG are early inhibitors of HCMV replication.
To identify what stage during virus replication was inhibited by CG, time of addition and time of removal experiments were performed. In the add-on group, compounds were added to infected HFFs at 0, 6, 12, 24, 36, and 48 hpi, and the luciferase was measured at 72 hpi. In the removal group, compounds were added immediately after virus infection and were subsequently removed after 6, 12, 24, 36, and 48 h; luciferase was measured at 72 hpi. Treatment with CG resulted in >90% inhibition of HCMV replication when added prior to or at 12 hpi. When CG were removed at 24 hpi, complete virus inhibition was already achieved, whereas GCV removal at that time point resulted in 60% virus inhibition (Fig: 2C and D). These data suggest that the effects of CG occur early in virus replication, prior to DNA replication.
CG inhibit HCMV IE gene expression.
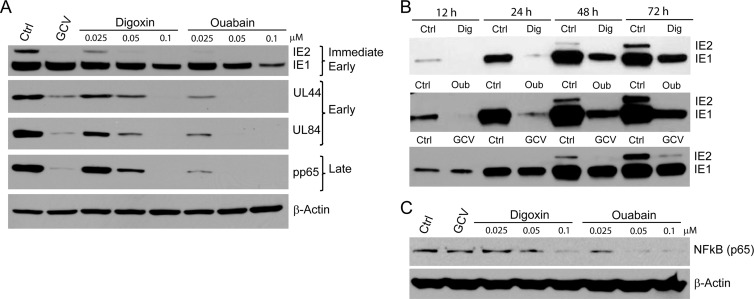
The expression of IE HCMV proteins IE1 and IE2 was quantified at 72 hpi (Fig. 3A, top panel). Digoxin and ouabain at 0.05 and 0.1 μM, respectively, decreased IE1 expression by ∼50 to 70% and ∼65 to 90%, whereas IE2 levels were undetected. The expression of the early genes UL84 and UL44 and the late gene pp65 was also undetectable at these concentrations of digoxin and ouabain (Fig. 3A). Ouabain was a more potent inhibitor of virus protein expression, similar to the luciferase assay data (Fig. 1).
Fig 3.
CG inhibit the expression of HCMV genes and NF-κB. (A) HFFs were infected and treated with the indicated concentrations of digoxin, ouabain, or GCV (10 μM). The expression of immediate-early (IE1 and IE2), early (UL84 and UL44), and late HCMV proteins (pp65) was determined in cell lysates after 72 h. (B) HFF cells were infected with HCMV, treated with digoxin (0.1 μM), ouabain (0.05 μM), or GCV (10 μM), and the expression levels of IE1 and IE2 proteins were determined at the indicated time points. (C) HFF cells were infected with HCMV and treated with DMSO (control), digoxin, or ouabain at the indicated concentrations. The concentration of GCV was 10 μM. The levels of NF-κB (p65 subunit) protein were determined after 12 h in total cell lysates. β-Actin was used as a sample loading control.
The effect of CG on IE1 and IE2 protein levels was determined at different times during virus replication (Fig. 3B). At all time points the levels of IE1 and IE2 were significantly reduced in treated HFFs (12 to 72 h). In contrast, at 12 and 24 hpi, there was no inhibition of IE1 by GCV; a slight decrease in IE1 expression was observed at 48 and 72 h. The levels of IE2 protein were significantly reduced by GCV at 48 and 72 h.
NF-κB expression is significantly downregulated by CG.
The expression of IE1 and IE2 proteins is regulated by the major IE (MIE) enhancer containing promoter, a region containing multiple binding sites for cellular transcription factors (TF) (17). Transactivation of the MIE after infection is required for generation of the MIE transcripts, viral replication and production of infectious virus. NF-κB is rapidly induced after HCMV infection and plays a critical role in the efficient transactivation of MIE, although other transcription factors such as the cyclic AMP response element-binding (CREB) and AP1 also activate the MIE (18, 26, 54). NF-κB levels were measured in infected and CG-treated HFFs. The levels of NF-κB (p65) protein were significantly reduced in total cell lysates at 12 hpi from HCMV-infected cells treated with digoxin at 0.1 μM and ouabain at 0.05 μM (Fig. 3C).
Combination of digoxin/ouabain and GCV is additive.
Since CG inhibited HCMV at an earlier time than GCV and were active against a GCV-resistant HCMV strain, the combination of the compounds was tested for additive/synergistic activity against HCMV replication. Using the Bliss independency model (21), the combinations of digoxin or ouabain and GCV resulted in an additive effect, suggesting independent mechanisms of action (Table 2).
Table 2.
Combination of digoxin or ouabain and GCVa
| Drug 1 |
Drug 2 |
Combination |
|||
|---|---|---|---|---|---|
| Drug (concn [μM]) | Fold inhibition | Drug (concn [μM]) | Fold inhibition | Theoretical fold inhibition | Actual fold inhibition |
| GCV (2.5) | 3.41 ± 0.02 | Digoxin (0.025) | 1.14 ± 0.04 | 3.89 ± 0.06 | 4.05 ± 0.01 |
| GCV (2.5) | 3.08 ± 0.03 | Ouabain (0.015) | 1.83 ± 0.01 | 5.64 ± 0.04 | 6.33 ± 0.01 |
Combinations of digoxin or ouabain and GCV were tested for their effect on HCMV inhibition. The expected combined effect of two drugs was calculated using the Bliss equation. Fold inhibition means ± the standard deviations were calculated as the reciprocal of the fraction unaffected at a particular drug concentration. The ratio of the actual to the theoretical fold inhibition is close to 1, suggesting that these combinations are additive. Mean values of triplicate determinations from three independent experiments are shown.
HCMV inhibition by CG is associated with changes in hERG expression.
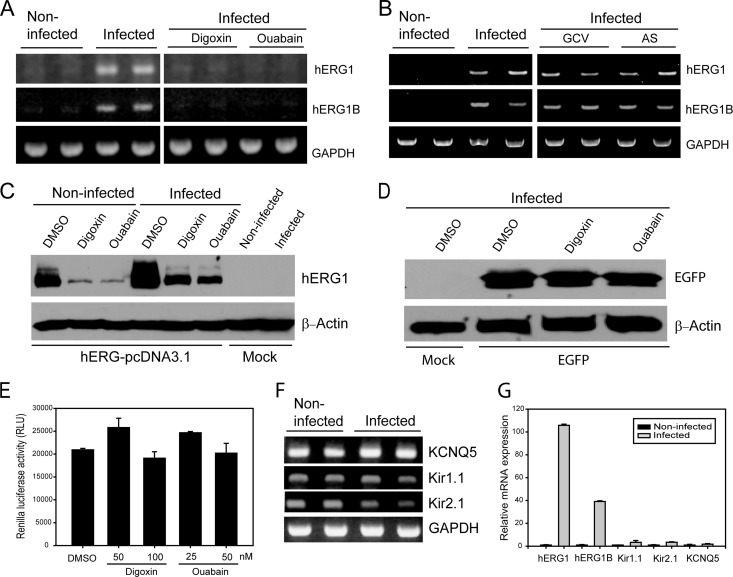
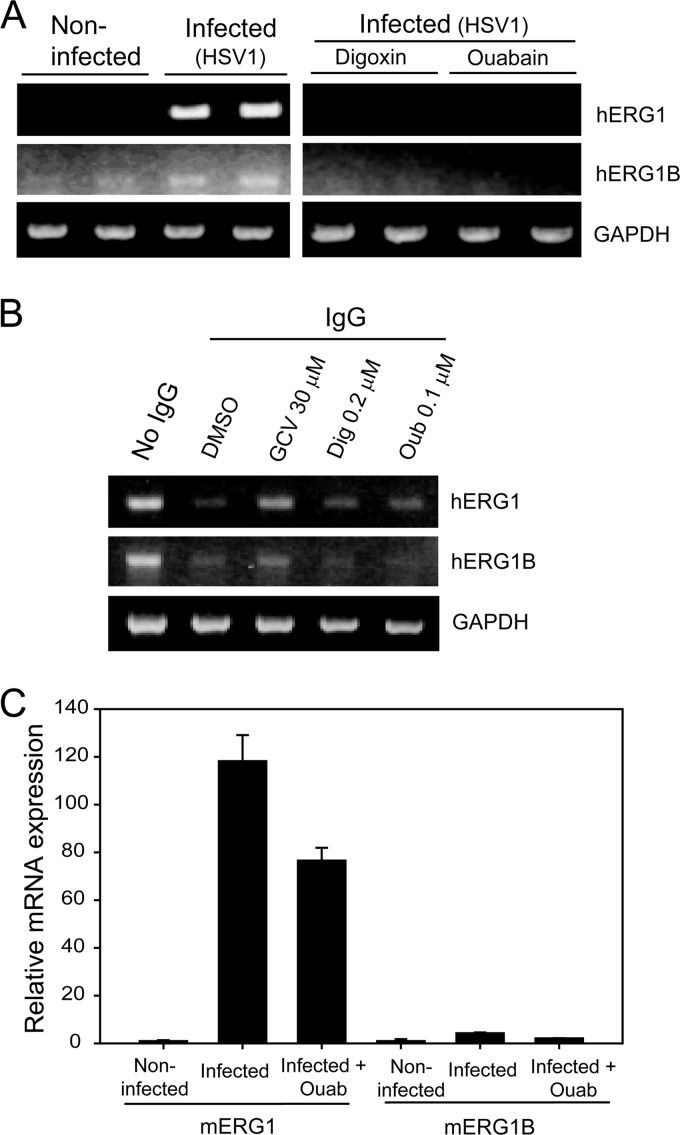
To assess whether the hERG gene contributed to the activity of CG in HCMV-infected cells, HFFs were infected with HCMV Towne and levels of hERG mRNA were measured (Fig. 4A). There are two N-terminal splice variants of hERG, including the full-length isoform 1 and the shorter isoform 1B (20). A significant increase in hERG1 and hERG1B transcripts was observed in HCMV-infected HFFs compared to mock-infected cells at 12 hpi (Fig. 4A). Both digoxin and ouabain completely inhibited the HCMV-mediated increase in hERG1 and hERG1B levels. The downregulation of hERG was specific to CG because mRNA levels of hERG in HCMV-infected cells treated with GCV were unchanged. In addition, treatment of HCMV-infected cells with artemisinin derivative (artesunate), an early inhibitor of HCMV replication, was not associated with changes in hERG mRNA (Fig. 4B) (1, 4).
Fig 4.
CG inhibit HCMV-mediated upregulation of hERG gene expression. (A and B) HFFs were infected with HCMV and treated with digoxin (0.1 μM) or ouabain (0.05 μM) (A) or GCV or artesunate (AS) (10 μM each) (B). After 12 h, the levels of hERG1, hERG1B, and GAPDH mRNA were measured by RT-PCR. (C) U373 cells were transiently transfected with hERG-pcDNA3.1 plasmid and at 6 hpi with HCMV. Immediately after infection cells were treated with DMSO, digoxin (0.1 μM), or ouabain (0.05 μM), and the levels of hERG1 protein and β-actin were determined by Western blotting after 24 h. (D) U373 cells were transiently transfected with EGFP-C plasmid and after 6 h were infected with HCMV. After infection, the cells were treated with DMSO, digoxin (0.1 μM), or ouabain (0.05 μM), and the levels of EGFP protein were determined by Western blotting after 24 h. (E) U373 cells were transfected with pRL-CMV plasmid and, after 6 h, treated with the indicated concentrations of digoxin and ouabain. The Renilla luciferase activity was measured in cell lysates after 24 h. (F) HFF cells were infected with HCMV, and the transcription of the potassium channels KCNQ5, Kir1.1, and Kir2.1 was determined by using RT-PCR at 12 hpi. (G) HFF cells were infected with HCMV, and the levels of hERG1, hERG1B, KCNQ5, Kir1.1, and Kir2.1 were measured by qRT-PCR. For each gene, the relative change in gene transcription between noninfected and infected cells was determined. Quantitative data represent mean values ± the SD of triplicate determinations from two independent experiments.
To determine the effects of CG on hERG protein expression in HCMV-infected cells, U373 cells were transiently transfected with hERG cDNA. At 6 h after transfection, the cells were infected and treated with digoxin (0.1 μM) or ouabain (0.05 μM). There was a significant increase in hERG1 protein expression in HCMV-infected-hERG-transfected cells compared to noninfected hERG-transfected-only cells at 24 h (Fig. 4C). Furthermore, treatment with CG significantly inhibited the HCMV-mediated hERG1 upregulation at the mRNA and protein level (Fig. 4A and C). The inhibition of hERG1 protein expression by CG in CMV-infected cells was not a result of inhibition of the CMV promoter in the hERG-pcDNA3.1 plasmid because the same experiment performed with an EGFP-C plasmid which expresses EGFP under the control of the same CMV promoter revealed no significant decrease in EGFP protein levels in DMSO- or CG-treated cells (Fig. 4D). In addition, transfection of U373 cells with a pRL-CMV reporter plasmid which expresses Renilla luciferase under the control of the CMV promoter, followed by treatment with digoxin, ouabain, or DMSO for 24 h revealed no significant changes in Renilla luciferase expression between mock (DMSO)-treated or CG-treated pRL-CMV-transfected cells.
To determine whether the effect of HCMV on hERG1 expression was specific to this channel, HFFs were infected with HCMV, and the expression of representative potassium channels (KCNQ5, Kir1.1, and Kir2.1) was measured at 12 hpi. These three channels were already expressed in noninfected HFFs, and their mRNAs did not change significantly upon HCMV infection (Fig. 4C). We quantified the changes in mRNA levels of hERG and the other representative potassium channels in noninfected and HCMV-infected HFFs by qRT-PCR. Although a significant increase was observed in hERG1 and hERG1B mRNA (100- and 40-fold, respectively), the changes observed in mRNA levels of KCNQ5, Kir1.1, and Kir2.1 in HCMV-infected cells compared to noninfected cells were <4-fold (Fig. 4G).
Downregulation of hERG transcription was also observed in HSV-1-infected HFFs treated with CG (Fig. 5A). In the case of EBV, a different pattern of hERG transcription was observed because endogenous hERG transcripts were already detected in the cancer Akata cells. Induction of lytic EBV replication was associated with significant downregulation of hERG (Fig. 5B). After lytic replication, the levels of hERG transcripts remained low in cells treated with CG but were restored in cells treated with GCV (Fig. 5B). Because both CG and GCV inhibited EBV lytic replication, these results suggest that hERG may play a role in the switch between the latent and lytic phases of EBV and that CG exert their inhibitory effects on EBV replication at least in part by maintaining the downregulation of hERG transcription.
Fig 5.
hERG expression in other herpesviruses. (A) HFF cells were infected with HSV-1 and then treated with digoxin (0.1 μM) or ouabain (0.05 μM). The mRNA levels of hERG1, hERG1B, and GAPDH were then determined at 12 hpi using RT-PCR. (B) Akata (EBV+) cells were induced by goat antihuman IgG, and the cells were treated with digoxin, ouabain, or GCV at the indicated concentrations. The transcription levels of hERG were determined by RT-PCR after 48 h. (C) MEFs were infected with MCMV, and the mRNA levels of mERG1, mERG1B, and GAPDH were measured at 12 hpi by using qRT-PCR. The concentration of ouabain used was 50 μM. Quantitative data represent mean values ± the SD of triplicate determinations from two independent experiments.
Because HCMV upregulated hERG transcription/translation and its expression levels were decreased at an early time point upon treatment with CG, we evaluated whether specific hERG inhibitors could inhibit HCMV replication. Dofetilide and E4031, potent and specific inhibitors of hERG channels, were used at concentrations ranging from 0.001 to 1 μM. These compounds did not inhibit HCMV replication (data not shown). Therefore, in HCMV-infected cells the effect of CG on hERG expression is indirect and involves a pathway downstream of the Na+,K+-ATPase.
CG do not inhibit MCMV replication.
To determine whether CG inhibit MCMV replication, a plaque reduction assay was performed. Concentrations of digoxin and ouabain as high as 100 and 50 μM, respectively, did not inhibit MCMV replication, whereas GCV (10 μM) completely inhibited MCMV (data not shown). The levels of mouse ERG (mERG) were quantified by RT-PCR in both MCMV-infected and MCMV-infected/CG-treated cells. There were nearly 120- and 4-fold increases in the mRNA levels of mERG1 and mERG1B, respectively, in MCMV-infected compared to uninfected MEF cells. However, treatment with 50 μM ouabain resulted in a <2-fold decrease in mERG1 and merg1B levels compared to infected-only cells (Fig. 5C). Thus, CG had no significant effect on the mERG levels in MCMV-infected cells.
DISCUSSION
The viral DNA polymerase inhibitors (GCV, foscarnet, and cidofovir) effectively suppress virus replication, but their prolonged use leads to significant side effects and the emergence of resistant mutants (19, 43, 46).
Compounds with specific effects on cellular pathways have been reported to inhibit HCMV replication. In general, these can be grouped into early inhibitors of HCMV replication, i.e., inhibitors of phosphoinositide 3-kinase (PI3-K) (23), p38 mitogen-activated protein kinase (MAPK) (22), and the tyrosine kinase inhibitor Gleevec (45), and others active at multiple steps of virus replication, including cellular cyclin-dependent kinase inhibitor, roscovitine (41), and leflunomide and its active metabolite FK778 (8).
CG have anticancer activities, and we now provide strong evidence for their antiviral activities. We show that these compounds are potent inhibitors of HCMV replication at nanomolar concentrations, similar to the concentrations that inhibit cancer cell growth (33). HSV-1 (alphaherpesvirus) and possibly EBV (gammaherpesvirus) were also inhibited by CG, suggesting the antiviral activities may involve cellular pathways or a viral protein shared by these herpesviruses. Because of the high sensitivity of the latently infected Akata cells to CG, the selectivity index of these compounds for EBV was less than that calculated for HCMV and HSV-1. Thus, inhibition of EBV replication requires further studies using other cell types.
Similar to the reported early inhibition of HSV-1 by CG (7), HCMV inhibition occurred early during virus replication. The expression of IE1 and IE2 proteins was decreased in the presence of CG, suggesting either an involvement of the MIE promoter and its activation by cellular transcription factors or a process that occurs after the transcription of the IE genes. The requirement for NF-κB in the transactivation of the MIE promoter has been described (3), and we show that CG significantly reduce the expression of NF-κB. We note that the MIE promoter has been shown to be activated by multiple transcription factors, and our data suggest that CG do not affect its transcription. Thus, the low levels of NF-κB protein measured at 12 hpi may represent an effect that occurs after the activation of the MIE.
How would CG inhibit HCMV? These compounds bind to the Na+,K+-ATPase that has an established role in ion transport. However, this pump has additional cellular activities, such as the regulation of cell growth and the expression of various genes (37). Binding of CG to the Na+,K+-ATPase triggers a signaling cascade (signalosome), resulting in activation of the protein tyrosine kinase Src, which transactivates epidermal growth factor receptor, Ras, and MAPKs. The diverse mechanisms of control of cell proliferation have been reviewed in detail (42). Some of the involved pathways are also used by HCMV for its efficient replication (53), and the fact that NF-κB expression was decreased in CG-treated cells supports this interpretation. Currently, there is no evidence to support direct inhibition of a virus-specific protein by CG. Proof that these compounds indeed target a specific viral protein will require demonstration that they can select for resistant viral mutants (16). These studies are ongoing. Alternatively, CG may target a cell protein or pathway(s) that are critical for the replication of herpesviruses.
Another mechanism that could link inhibition of cancer cells and HCMV by CG involves an interaction between the Na+,K+-ATPase and other ion channels. We show that HCMV significantly upregulate hERG transcription as early as 12 hpi, while CG downregulate its transcription. qRT-PCR showed that the transcription of three other potassium channels was unchanged during HCMV infection. The hERG gene has been associated with multiple cancers and appears to be important in cancer progression in vivo. Its role in virus infections has not been studied (2, 35, 36). Although MCMV infection resulted in the upregulation of mERG, treatment with CG did not inhibit virus replication, and mERG levels were unchanged during infection and treatment with CG. Interspecies differences in the effects of CG have been noted in studies using a variety of cells (12, 40). Transformation of human lymphocytes in vitro by phytohemagglutinin was sensitive to ouabain, but rat cultures were relatively insensitive (38). In addition, tumor cell sensitivity to CG was reported to be species dependent. Although nontoxic to rodent-derived tumor cell lines, CG are potent inhibitors of human tumor cells (39). These differences in activity may be due to changes in the structure of the Na+,K+-ATPase among species.
At therapeutic concentrations (0.0006 to 0.0015 μM), digoxin is unlikely to inhibit HCMV replication based on the in vitro concentrations reported here. However, work has shown that digitoxin may have anticancer effects at concentrations commonly found in cardiac patients (0.02 to 0.033 μM) (14, 29). Based on our in vitro concentrations of HCMV inhibition by digitoxin (EC50 = 0.016 μM ± 0.00), digitoxin may have inhibitory effects on HCMV in vivo. Thus, studies of additional CG with potential clinical application are needed.
In conclusion, CG are potent inhibitors of HCMV replication and IE gene expression. Their effects correlate with the transcription and translation of hERG. Future studies may characterize the signaling and chaperons involved in this interaction.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants 1R01AI093701 and KO8 AI074907 to R.A.-B.
We thank Gordon Sandford for providing the MCMV Smith strain, Diane Hayward and Rengfeng Li for the EBV Akata strain, and David A. Leib, Dartmouth Medical School, for the HSV-1 KOS/Dlux/oriS. We also thank Min Li for the hERG inhibitors and the hERG plasmid.
Footnotes
Published ahead of print 9 July 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Arav-Boger R, et al. 2010. Artemisinin-derived dimers have greatly improved anti-cytomegalovirus activity compared to artemisinin monomers. PLoS One 5:e10370 doi:10.1371/journal.pone.0010370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bianchi L, et al. 1998. hERG encodes a K+ current highly conserved in tumors of different histogenesis: a selective advantage for cancer cells? Cancer Res. 58:815–822 [PubMed] [Google Scholar]
- 3. Cherrington JM, Mocarski ES. 1989. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chou S, et al. 2011. The unique antiviral activity of artesunate is broadly effective against human cytomegaloviruses including therapy-resistant mutants. Antivir. Res. 92:364–368 [DOI] [PubMed] [Google Scholar]
- 5. Chou SW. 2001. Cytomegalovirus drug resistance and clinical implications. Transpl. Infect. Dis. 3(Suppl 2):20–24 [DOI] [PubMed] [Google Scholar]
- 6. Demmler GJ. 1991. Infectious Diseases Society of America and Centers for Disease Control: summary of a workshop on surveillance for congenital cytomegalovirus disease. Rev. Infect. Dis. 13:315–329 [DOI] [PubMed] [Google Scholar]
- 7. Dodson AW, Taylor TJ, Knipe DM, Coen DM. 2007. Inhibitors of the sodium potassium ATPase that impair herpes simplex virus replication identified via a chemical screening approach. Virology 366:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evers DL, Wang X, Huong SM, Andreoni KA, Huang ES. 2005. Inhibition of human cytomegalovirus signaling and replication by the immunosuppressant FK778. Antivir. Res. 65:1–12 [DOI] [PubMed] [Google Scholar]
- 9. Ficker E, Dennis AT, Wang L, Brown AM. 2003. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ. Res. 92:e87–e100 [DOI] [PubMed] [Google Scholar]
- 10. Goldner T, et al. 2011. The novel anti-cytomegalovirus compound AIC246 inhibits HCMV replication through a specific antiviral mechanism that involves the viral terminase. J. Virol. 85:10884–10893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffiths PD, Clark DA, Emery VC. 2000. Betaherpesviruses in transplant recipients. J. Antimicrob. Chemother. 45 (Suppl T3):29–34 [DOI] [PubMed] [Google Scholar]
- 12. Gupta RS, Chopra A, Stetsko DK. 1986. Cellular basis for the species differences in sensitivity to cardiac glycosides (digitalis). J. Cell Physiol. 127:197–206 [DOI] [PubMed] [Google Scholar]
- 13. Hartley C, Hartley M, Pardoe I, Knight A. 2006. Ionic contra-viral therapy (ICVT); a new approach to the treatment of DNA virus infections. Arch. Virol. 151:2495–2501 [DOI] [PubMed] [Google Scholar]
- 14. Haux J, Klepp O, Spigset O, Tretli S. 2001. Digitoxin medication and cancer: case control and internal dose-response studies. BMC Cancer 1:11 doi:10.1186/1471-2407-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He R, et al. 2011. Recombinant luciferase-expressing human cytomegalovirus (CMV) for evaluation of CMV inhibitors. Virol. J. 8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herrmann EC, Jr, Herrmann JA. 1977. A working hypothesis: virus resistance development as an indicator of specific antiviral activity. Ann. N. Y. Acad. Sci. 284:632–637 [DOI] [PubMed] [Google Scholar]
- 17. Isomura H, Stinski MF. 2003. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J. Virol. 77:3602–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isomura H, et al. 2005. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 79:9597–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jabs DA, Martin BK, Forman MS. 2010. Mortality associated with resistant cytomegalovirus among patients with cytomegalovirus retinitis and AIDS. Ophthalmology 117:128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jenke M, et al. 2003. C-terminal domains implicated in the functional surface expression of potassium channels. EMBO J. 22:395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jilek BL, et al. 2012. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat. Med. 18:446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson RA, Huong SM, Huang ES. 2000. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J. Virol. 74:1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson RA, Wang X, Ma XL, Huong SM, Huang ES. 2001. Human cytomegalovirus upregulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J. Virol. 75:6022–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kovacs A, et al. 1999. Cytomegalovirus infection and HIV-1 disease progression in infants born to HIV-1-infected women. N. Engl. J. Med. 341:77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langenhan JM, Peters NR, Guzei IA, Hoffmann FM, Thorson JS. 2005. Enhancing the anticancer properties of cardiac glycosides by neoglycorandomization. Proc. Natl. Acad. Sci. U. S. A. 102:12305–12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lashmit P, Wang S, Li H, Isomura H, Stinski MF. 2009. The CREB site in the proximal enhancer is critical for cooperative interaction with the other transcription factor binding sites to enhance transcription of the major intermediate-early genes in human cytomegalovirus-infected cells. J. Virol. 83:8893–8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li R, et al. 2011. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Limaye AP, et al. 2008. Cytomegalovirus reactivation in critically ill immunocompetent patients. JAMA 300:413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Lazaro M, et al. 2005. Digitoxin inhibits the growth of cancer cell lines at concentrations commonly found in cardiac patients. J. Nat. Prod. 68:1642–1645 [DOI] [PubMed] [Google Scholar]
- 30. Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mijatovic T, et al. 2007. Cardiotonic steroids on the road to anti-cancer therapy. Biochim. Biophys. Acta 1776:32–57 [DOI] [PubMed] [Google Scholar]
- 32. Mitchell DA, et al. 2008. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro. Oncol. 10:10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newman RA, Yang P, Pawlus AD, Block KI. 2008. Cardiac glycosides as novel cancer therapeutic agents. Mol. Interv. 8:36–49 [DOI] [PubMed] [Google Scholar]
- 34. Osawa R, Singh N. 2009. Cytomegalovirus infection in critically ill patients: a systematic review. Crit. Care. 13:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pardo LA, Contreras-Jurado C, Zientkowska M, Alves F, Stuhmer W. 2005. Role of voltage-gated potassium channels in cancer. J. Membr. Biol. 205:115–124 [DOI] [PubMed] [Google Scholar]
- 36. Pillozzi S, et al. 2002. HERG potassium channels are constitutively expressed in primary human acute myeloid leukemias and regulate cell proliferation of normal and leukemic hemopoietic progenitors. Leukemia 16:1791–1798 [DOI] [PubMed] [Google Scholar]
- 37. Prassas I, Diamandis EP. 2008. Novel therapeutic applications of cardiac glycosides. Nat. Rev. Drug Discov. 7:926–935 [DOI] [PubMed] [Google Scholar]
- 38. Quastel MR, Vogelfanger IJ. 1971. Interspecies differences in the inhibitory effect of ouabain on lymphocyte transformation. Cell. Immunol. 2:504–507 [DOI] [PubMed] [Google Scholar]
- 39. Raghavendra PB, Sreenivasan Y, Manna SK. 2007. Oleandrin induces apoptosis in human, but not in murine cells: dephosphorylation of Akt, expression of FasL, and alteration of membrane fluidity. Mol. Immunol. 44:2292–2302 [DOI] [PubMed] [Google Scholar]
- 40. Repke K, Est M, Portius HJ. 1965. On the cause of species differences in digitalis sensitivity. Biochem. Pharmacol. 14:1785–1802 [DOI] [PubMed] [Google Scholar]
- 41. Sanchez V, et al. 2004. Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J. Virol. 78:11219–11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schoner W, Scheiner-Bobis G. 2007. Endogenous and exogenous cardiac glycosides: their roles in hypertension, salt metabolism, and cell growth. Am. J. Physiol. Cell Physiol. 293:C509–C536 [DOI] [PubMed] [Google Scholar]
- 43. Schreiber A, et al. 2009. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 10:191–209 [DOI] [PubMed] [Google Scholar]
- 44. Shen L, et al. 2008. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat. Med. 14:762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soroceanu L, Akhavan A, Cobbs CS. 2008. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature 455:391–395 [DOI] [PubMed] [Google Scholar]
- 46. Steininger C. 2007. Novel therapies for cytomegalovirus disease. Recent Pat. Antiinfect. Drug Discov. 2:53–72 [DOI] [PubMed] [Google Scholar]
- 47. Stenkvist B. 1999. Is digitalis a therapy for breast carcinoma? Oncol. Rep. 6:493–496 [PubMed] [Google Scholar]
- 48. Summers BC, Margolis TP, Leib DA. 2001. Herpes simplex virus type 1 corneal infection results in periocular disease by zosteriform spread. J. Virol. 75:5069–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanaka Y, et al. 2002. Monitoring cytomegalovirus infection by antigenemia assay and two distinct plasma real-time PCR methods after hematopoietic stem cell transplantation. Bone Marrow Transplant. 30:315–319 [DOI] [PubMed] [Google Scholar]
- 50. Wang L, Wible BA, Wan X, Ficker E. 2007. Cardiac glycosides as novel inhibitors of human ether-a-go-go-related gene channel trafficking. J. Pharmacol. Exp. Ther. 320:525–534 [DOI] [PubMed] [Google Scholar]
- 51. Williams SL, et al. 2003. In vitro activities of benzimidazole D- and L-ribonucleosides against herpesviruses. Antimicrob. Agents Chemother. 47:2186–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie Z, Cai T. 2003. Na+-K+–ATPase-mediated signal transduction: from protein interaction to cellular function. Mol. Interv. 3:157–168 [DOI] [PubMed] [Google Scholar]
- 53. Yurochko AD. 2008. Human cytomegalovirus modulation of signal transduction. Curr. Top. Microbiol. Immunol. 325:205–220 [DOI] [PubMed] [Google Scholar]
- 54. Yurochko AD, et al. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.