Abstract
Among the C1A cysteine proteases, the plant cathepsin F-like group has been poorly studied. This paper describes the molecular and functional characterization of the HvPap-1 cathepsin F-like protein from barley. This peptidase is N-glycosylated and has to be processed to become active by its own propeptide being an important modulator of the peptidase activity. The expression pattern of its mRNA and protein suggest that it is involved in different proteolytic processes in the barley plant. HvPap-1 peptidase has been purified in Escherichia coli and the recombinant protein is able to degrade different substrates, including barley grain proteins (hordeins, albumins, and globulins) stored in the barley endosperm. It has been localized in protein bodies and vesicles of the embryo and it is induced in aleurones by gibberellin treatment. These three features support the implication of HvPap-1 in storage protein mobilization during grain germination. In addition, a complex regulation exerted by the barley cystatins, which are cysteine protease inhibitors, and by its own propeptide, is also described
Key words: Barley grain, cathepsin F-like peptidase, cystatin, cysteine protease, grain germination, propeptide, protein mobilization, proteolysis.
Introduction
Plant proteolysis is a complex process that involves broad metabolic networks, different subcellular compartments, and various types of peptidases, mainly cysteine, serine, aspartic, and metallo-proteases (van der Hoorn, 2008). Protease activities are regulated at the transcriptional level by differential gene expression, and at the protein level by the activation of inactive zymogens and by the binding of specific inhibitors and cofactors. Among the >800 proteases encoded by a plant genome, ~140 correspond to cysteine proteases (CysProts) that belong to 15 families distributed in five clans as classified in the MEROPS peptidase database (Rawlings et al., 2012). The most analysed CysProts are the papain-like proteases, known as C1A (family C1, clan CA). This group of proteases is extensively present in land plants, with members ranking from 32 in Arabidopsis to 45 in rice (Martinez and Diaz, 2008). C1A CysProts commonly contain three disulphide bonds and their chain is folded to form a globular protein with two interacting domains delimiting a cleft at the surface where substrates can be bound (Kamphuis et al., 1985). The evolutionarily highly conserved catalytic mechanism of these peptidases involves three amino acids, cysteine, histidine, and asparagine, in the catalytic triad, and a glutamine which seems to be essential for maintaining an active enzyme conformation. These proteins are synthesized as inactive precursors, which comprise an N-terminal signal peptide, a 130–250 residues prosequence, upstream of the mature protein. Activation takes place by limited intra- or intermolecular proteolysis cleaving off an inhibitory propeptide (Wiederanders et al., 2003). C1A CysProts from plants are synthesized as pre-proteins, processed either autocatalytically or with the aid of processing enzymes, transported via the trans-Golgi network, and finally either stored in the vacuoles or externally secreted (Grudkowska and Zagdanska, 2004).
Based on their similarity to mammalian proteases, the C1A proteases from plants have been grouped as cathepsin L-, B-, H-, or F-like (Martinez and Diaz, 2008). B-like proteins share a sequence similar to human cathepsin-B, which is dissimilar to other C1A CysProts (Karrer et al., 1993). H- and L-like proteins, closer in amino acid sequence, conserve a common motif in their prosequence, the ERFNIN signature, but are evolutionarily separated in phylogenetic analyses (Martinez and Diaz, 2008). F-like proteases maintain an ERFNAQ motif in their prosequence typical of this kind of cathepsins, and phylogenetic analysis postulated them as a new subgroup of cathepsins (Wex et al., 1999). In spite of the high number of identified genes encoding C1A CysProts, information about their roles is unknown or still fragmentary (Martinez et al., 2012). They participate in protein degradation during senescence and abscission processes, programmed cell death, fruit ripening, and the accumulation and mobilization of storage proteins in seeds and tubers (Grudkowska and Zagdanska, 2004; Martinez et al., 2007, 2009; van der Hoorn, 2008; Shi and Xu, 2009; Weeda et al., 2009; Esteban-Garcia et al., 2010; Parrott et al., 2010). Moreover, the expression of CysProt-encoding genes is enhanced under various environmental stresses, which triggers reorganization of metabolism, remodelling of cell protein components, degradation of damaged or unnecessary proteins, and nutrient remobilization. They also play an essential role in local and systemic defence responses against pathogens and pests (Pechan et al., 2000; Shindo and Van der Hoorn, 2008; McLellan et al., 2009; Kaschani and Van der Hoorn, 2011; Shindo et al., 2012).
Regarding their roles in the grain, C1A CysProts are described as one of the most abundant group of proteases responsible for the degradation and mobilization of storage proteins in seeds (Fischer et al., 2000; Grudkowska and Zagdanska, 2004; Sreenivasulu et al., 2008). Grains act as a store of starch, proteins, and lipids that are used during the germination process. Events occurring in this period are crucial for the survival of the seedling until photosynthesis is fully established. Hydrolysis of reserves is a key point, which is accomplished by several enzymes, such as amylases and proteases. These hydrolases are either stored during seed maturation or newly synthesized during early germination. The products of this degradation process are absorbed by the scutellum and utilized for seedling development.
Barley grain is the model for germination research in monocots. Most analyses have been focused at the transcriptional level. A complex network of genes encoding transcriptional factors in response to phytohormones is involved in the control of hydrolase expression at the beginning of germination (Gubler et al., 1999; Moreno-Risueno et al., 2007; Sreenivasulu et al., 2008). In contrast, there are few data related to post-germination processes where hydrolases mobilize storage compounds. In barley, there are at least 32 C1A CysProts, which belong to the L-, H-, F-, and B-like cathepsin groups (Martinez and Diaz, 2008). A previous analysis reported that 27 CysProts are among the 42 proteases that are involved in the germination of barley grain (Zhang and Jones, 1995). More recently, Sreenivasulu et al. (2008) have published a complete transcriptome analysis of barley grain germination in two tissue fractions (starchy endosperm/aleurone and embryo/scutellum) and have shown the induction of a high number of CysProt genes during germination, most of them being mediated by gibberellins (GAs). Until now, few of the barley C1A proteases expressed in grain tissues have been characterized. Among them, several cathepsin L-like proteases were present in the scutellar epithelium and the aleurone layer, and were secreted to the endosperm upon germination in response to GA (Koehler and Ho, 1990; Mikkonen et al., 1996; Martinez et al., 2009). Moreover, a cathepsin H-like protein isolated from GA-induced aleurone cells was targeted to vacuoles, and a cathepsin B-like protein was expressed in the aleurone and induced by GA treatment (Holwerda and Rogers, 1992; Martinez et al., 2003). In addition, there are only limited data on the effects of the inhibitors required for their regulation. The most well known proteinaceous inhibitors of C1A CysProts are cystatins, which are extensively present in plants, and their inhibitory properties have been studied in depth (Benchabane et al., 2010). Recently, the participation of some cathepsin L-like proteins and cystatins from barley as partners in hordein mobilization during seed germination has been reported (Martinez et al., 2009). Their subcellular location in the endoplasmic reticulum (ER) and Golgi system has been detected as part of the secretory route through which the proteases transit to reach the protein bodies where the target proteins are stored. Barley CysProts and their inhibitors (cystatins) co-localize in the endomembrane system and also interact in vivo in the plant cell, as revealed by bimolecular fluorescent complementation (BiFC) experiments. The functional relationship between cathepsin L-like proteins and cystatins has been shown through their implication as counterparts in the mobilization of hordeins stored in the seed.
This study covers the molecular characterization of a new cathepsin F-like CysProt, HvPap-1, from barley. The expression of HvPap-1 in several barley tissues, its subcellular location in embryos, and its response in aleurone layers to GA are shown. The functional role of HvPap-1 in mobilizing storage proteins, mainly hordeins, regulated either by specific inhibitors (cystatins) or by its own propeptide is also discussed.
Materials and methods
Real-time quantitative PCR analysis
For real-time quantitative PCR (qRT-PCR) studies, grains of barley (Hordeum vulgare) cv. Bomi were germinated at 22 ºC in the dark for 7 d and used to collect samples of leaves and roots. Developing endosperm at 10–22 days after flowering (daf) and immature embryo (18 daf) were prepared from kernels of plants grown in a greenhouse at 18 ºC under an 18 h light/6 h dark photoperiod. Mature embryos were isolated from dry barley Bomi grains. Germinating embryos were obtained from imbibed grains at 8, 16, 24, and 48 hours after imbibition (hai) at 22 ºC in the dark. All samples were frozen in liquid N2 and stored at –80 ºC until used for RNA–protein extraction. Isolated aleurone layers from embryoless barley grains (cv. Himalaya) were prepared (Moreno-Risueno et al., 2007), incubated in the presence or absence of 1 µM GA3 for 8, 24, and 48 h in a calcium succinate buffer (20 mM Na-succinate, 20 mM CaCl2, pH 5.2), and then frozen.
Total RNA was extracted from frozen samples by the phenol/chloroform method, followed by precipitation with 3 M LiCl (Lagrimini et al., 1987) and digestion with DNase. cDNAs were synthesized from 1 µg of RNA using the High Reverse Transcription kit (Applied Biosystems) following the manufacturer’s instructions. qRT-PCR analyses were performed for duplicated samples by means of a CFX96 Real-time system (BioRad) using a SYBR Green detection system. Quantification was standardized to barley Actin2 mRNA levels. The primers used for PCR amplification were as follows: Actin2 forward, 5'-TCCACCGGAGAGGAAGTACAGT-3'; Actin2 reverse, 5'-AATGTGCTCAGAGATGCAAGGA-3'; HvPap-1 forward, 5'-TCCTGGAGTCGATCTTGGTTTC-3'; and HvPap-1 reverse, 5'-CAAGCATACTGTTGCGGCTTC-3'.
Protease sequence and phylogenetic analysis
The nucleotide and amino acid sequences of HvPap-1 were extracted from the NCBI GenBank (accession no. BN000093). Signal peptide analysis was performed using the SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP) program (Bendtsen et al., 2004). Prediction of N-glycosylation sites was carried out by the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc). The three-dimensional structure of the barley HvPap-1 protein was modelled by the automated SWISS-MODEL program (Bordoli et al., 2009). The known crystal structure of the human cathepsin-F (PDB identifier 1M6D) was used to construct the homology-based models. Structure analysis was performed using the RasMol 2.7 program (Sayle and Milner-White, 1995).
BlastP searches for cathepsin F-like cysteine peptidases were performed in the comparative genomics database Phytozome v7.0 (http://www.phytozome.net). Phytozome provides access to 23 sequenced and annotated green plant genomes. Searches were made in the current releases for: Arabidopsis thaliana v9.0; Populus trichocarpa v2.2; Ricinus communis v0.1; Brachypodium distachyon v1.0; Oryza sativa v6.0; and Sorghum bicolor v1.0. Blast searches were made using the amino acid sequence of the HvPap-1 protein. Proteins that conserve the specific ERFNAQ motif for cathepsin-F-like proteases were selected. Cathepsin F-like proteins from H. vulgare were previously described (Martinez and Diaz, 2008). Information about gene models for all these proteins is compiled in Supplementary Fig. S1 available at JXB online. Phylogenetic analysis was carried out using the program MEGA version 5.0 (http://www.megasoftware.net) (Tamura et al., 2011). Alignments of the amino acid sequences were performed using the default parameters of MUSCLE (Edgar, 2004). The displayed cathepsin-F proteases tree was performed by means of the Neighbor–Joining method. Bootstrapping was used as a statistical test. The tree was rooted using as outgroup protein a cathepsin F-like sequence belonging to the spikemoss Selaginella moellendorffii.
Expression, purification, and activation of recombinant proteins from E.coli
The cDNA fragment spanning the whole HvPap-1 open reading frame (ORF), apart from its signal peptide sequences, was amplified by PCR and inserted in-frame into the expression vector pRSETB (Invitrogen). As described by Bethune et al. (2006), bacterial cells were grown at 37 °C to an OD550 of ~0.5, induced with 0.25 mM IPTG (isopropyl β-d-thiogalactopyranoside) for 20 h, harvested, and processed. Constructs encoding the protease with both N- and C-terminal His6 tags were purified using an Ni-NTA agarose column (Qiagen) following the manufacturer’s instructions. After dialysis, the purification process was checked by SDS–PAGE. The final protein concentration was quantified by the BioRad kit with bovine serum albumin (BSA) as standard. Activation of the HvPap-1 protein was checked by diluting the protease by a ratio of 1:4 (v:v) in a buffer containing 100 mM sodium acetate at pH 4.0, or by diluting the protease to 1:4 (v:v) in a buffer containing 100 mM TRIS-HCl pH 8.0, 5 mM EDTA, 15% glycerol (v/v), and 2 mM β-mercaptoethanol plus pepsin at a concentration of 0.6 mg ml–1, or by a combination of both treatments.
For propeptides, the cDNA fragment spanning the whole HvPap-1 propeptide (HvPap-1pro, A25–G141) or a form truncated in the N-terminus (HvPap-1pro*, D44–G141) was amplified by PCR and inserted in-frame into the expression vector pRSETB. Cystatins (HvCPI-1 to HvCPI-13) and propeptides (HvPap-1pro and HvPap-1pro*) were expressed and purified as previously described (Martinez et al., 2009). pRSETB expression plasmids containing the cystatin or propeptide sequences were introduced into E. coli BL21 CodonPlus (Stratagene). Bacterial cells were grown at 37 ºC to an OD550 of ~0.5–0.6 and induced with 1 mM IPTG for 2 h, harvested, and processed. The fusion proteins with the histidine tail were purified using a His-Bind Resin (Novagen) following the manufacturer’s instructions. The purification process was checked by SDS–PAGE. The final protein concentration was quantified by the BioRad kit with BSA as standard.
Enzyme activity assays
The enzymatic activity of recombinant HvPap-1 was routinely determined using the fluorogenic substrates Z-FR-AMC (N-carbobenzoxyloxy-Phe-Arg-7-amido-4-methylcoumarin) and Z-RR-AMC (N-carbobenzoxyloxy-Arg-Arg-7-amido-4-methylcoumarin) at 25 µM concentration and Bz-FVR-AMC (benzoyl-Phe-Val-Arg-7-amido-4-methylcoumarin) at 100 µM, susceptible to degradation by cathepsin L-, B-, and H-like proteases, respectively. Mixtures of proteases and substrates were incubated in 100 mM sodium phosphate pH 6.0 buffer, containing 10 mM cysteine, 1 mM EDTA, and 0.01% (v/v) Brij35 for 3 h at 30 ºC. Emitted fluorescence was measured with a 365 nm excitation wavelength filter and a 465 nm emission wavelength filter.
To determine the effect of pH, reactions were carried out in 100 mM sodium citrate buffer (pH 3–3.5), 100 mM sodium acetate buffer (pH 4–5.5), 100 mM sodium phosphate buffer (pH 6–7), and 100 mM TRIS (pH 7.5–9), supplemented with 10 mM l-cysteine, 10 mM EDTA, and 0.01% (v/v) Brij35 using the Z-FR-AMC substrate, for 3 h. The effect of the temperature was assayed in 100 mM sodium phosphate buffer pH 6.0, for 3 h. Triplicate assays were performed for determination of each value and the average was calculated. Blanks were used to account for the spontaneous breakdown of substrates. Emitted fluorescence was measured with a 365 nm excitation wavelength and a 465 nm emission wavelength.
Inhibitory activity of propeptides and cystatins against HvPap-1 protease
The recombinant propeptides (HvPap-1pro and HvPap-1pro*) were assayed against the recombinant HvPap-1 protease, both purified from E. coli cultures. The recombinant barley cystatins (Martinez et al., 2009) were also included for inhibitory experiments against HvPap-1. Briefly, different concentrations of propeptides or cystatins plus 300 ng of the protease were incubated in a buffer containing 100 mM sodium phosphate pH 6.0, 10 mM l-cysteine, 10 mM EDTA, and 0.01% (v/v) Brij35 at room temperature for 10 min. Then, the Z-FR-AMC substrate was added and the reactions were incubated for 1 h at 30 ºC. Emitted fluorescence was measured as above. The system was calibrated with known amounts of AMC hydrolysis product in a standard reaction mixture. All assays were carried out in triplicate and blanks were used to account for the spontaneous breakdown of substrates. As negative control, proteins from E. coli transformed with the empty expression vector were used. The type of inhibition was determined from Lineweaver–Burk plots (1/V versus 1/[S]). K i values were calculated by non-linear regression using the GraFit program (Leatherbarrow, 2009).
Western blot analysis
Plant protein extracts were obtained from frozen leaf, root, embryo, and endosperm barley tissues at the same developmental stages used for RNA preparation for the qRT-PCR analysis. Samples were ground and resuspended in a 50 mM sodium phosphate buffer, pH 6.0, containing 0.15 M sodium chloride and 2 mM EDTA. The suspension was centrifuged at 10 000 rpm for 10 min at 4 ºC. The protein content of the supernatant was quantified by the BioRad kit, using BSA as standard. After separation on an SDS–polyacrylamide gel, proteins were electrotransferred onto a nitrocellulose membrane (GE Healthcare). Immunoblotting was performed with anti-HvPap-1 peptide polyclonal antibody (HvPap-1-IgG) whose sequence is indicated in Fig. 1B. The antibody was produced in rabbits by Pineda Antibody Services (Berlin, Germany). Optimal dilution of the antibody at 1:5000 (v:v) in antisera buffer [0.05% (w/v) skim milk, 0.1% (v/v) 10× BS, and 0.001% Tween-20] was used. Peroxidase-conjugated anti-rabbit IgG (Sigma) diluted at 1:10 000 (v:v) was used as secondary antibody for detection with ECL Plus (GE Healthcare).
Fig. 1.
(A) Expression of the HvPap-1 gene in aleurone layers after 48 h of incubation in the presence or absence of 1 μM GA, determined by qRT-PCR. Values are expressed as relative mRNA levels of barley protease and standardized using the barley Actin2 mRNA content. (B) Nucleotide and deduced amino acid sequences of the cDNA encoding the cathepsin F-like HvPap-1 peptidase. The dark and light grey boxes represent the signal peptide and the propeptide sequences, respectively. The active site residues C166, H308, and N335, characteristic of C1A CysProts, are boxed. The conserved motif EX3RX3FX2NX3AX3Q specific for cathepsin F-like proteases is also boxed. The GXN/TXFXD and the GCNGG like-motifs, common to all CysProts, are double underlined. The consensus QVVGG sequence present in cystatins and responsible for the protease–inhibitor interaction is marked by a dotted box. Cysteine residues are circled and the stop codon is indicated by an asterisk. The predicted N-glycosylation motif NYS is underlined by a dotted line. The horizontal double arrowhead indicates the amino acid sequence used for specific antibody production (HvPap-1-IgG). (C) Ribbon plots of HvPap-1 and human cathepsin-F. The three-dimensional structure of HvPap-1 was predicted using the automated SWISS-MODEL program with the known human cathepsin-F structure as template (1M6D). The figure was prepared with RasMol 2.7. The active site residues of HvPap-1 are coloured in green. (This figure is available in colour at JXB online.)
Glycosylation assays
To isolate glycoproteins from barley leaves, 2 g of tissue were blended in 20 ml of extraction buffer (20 mM TRIS-HCl pH 7.4 containing 1 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, and 0.15 M NaCl) at 4 ºC. The crude extract was centrifuged at 15 000 rpm for 30 min at 4 ºC and the supernatant was precipitated with ammonium sulphate at 90% (w/v) saturation. The precipitate was collected after centrifugation at 15 000 rpm for 30 min, redissolved in 1.5 ml of deionized water, and dialysed against deionized water to remove salts. The desalted crude protein was concentrated using a freeze drier and dissolved in the extraction buffer to be applied to a concanavalin A (Con A)–Sepharose 4B Column (GE Healthcare). The chromatography was performed essentially as previously described (Boonmee et al., 2011). The eluted protein fraction was concentrated and desalted using centrifugal filters of 10K (Amicon ultra, Millipore). Western blot assays were performed as indicated above using the HvPap-1-IgG and the anti-N-glycosylation (N-Glyc-IgG) antibodies. The N-Glyc-IgG antibody, donated by Dr Diaz-Perales (CBGP-UPM-INIA, Madrid, Spain), was produced in rabbits.
Additionally, immunoprecipitation of native HvPap-1 was performed. Aliquots of protein extracts (20 µg) were prepared as indicated above from 24 h germinating embryos and 7-day-old barley leaves, and were incubated with polyclonal anti-HvPap-1 antibody (HvPap-1-IgG) for 1 h at room temperature. After the incubation, 4% (w/v) protein A–Sepharose was added and vortexed briefly (5 min intervals) for 15 min. The beaded immunocomplexes were sedimented by centrifugation at 12 000 rpm for 5 min, washed once with buffer [0.5 M TRIS-HCl, pH 8.0, 1.5 M NaCl, and 1% (v/v) Triton X-100], and twice with 0.1 M HEPES-NaOH, pH 7.1. Pellets were dissolved in SDS sample buffer [50 mM TRIS-HCl, pH 8.8, 20% (v/v) glycerol, 1% (w/v) SDS, 14 mM β-mercaptoethanol, and 0.05% (w/v) bromophenol blue], heated for 10 min at 90 °C, and centrifuged for 5 min at 12 000 rpm at room temperature. For immunoblot assays, membranes were first incubated with HvPap-1-IgG antibody and then were stripped using a stripping buffer [glycine-HCl 25 mM, pH 2.0 containing 1% (w/v) SDS]. Finally they were incubated with the N-Glyc-IgG antibody, after checking that membranes were clean.
Fractionation of albumins, globulins, and hordeins from barley grain
Albumins and globulins were sequentially extracted from barley (cv. Bomi) grains (Shi and Xu, 2009). Dry barley grains were completely crushed in a mortar, resuspended in distilled water, and continuously stirred for 12 h at 4 ºC. Soluble and insoluble fractions were separated by centrifugation at 8000 rpm for 30 min at 4 ºC. The supernatant, enriched in albumins, was isolated. Globulins were extracted from the pellet by adding 5% (w/v) NaCl in distilled water at 4 ºC. After a second centrifugation step at 8000 rpm for 30 min, the recovered soluble fraction was enriched in globulins. Hordeins were extracted from dry barley grains after incubation in a buffer containing 55% (v/v) 2-propanol and 1% (v/v) 2-mercaptoethanol, for 1 h at 60 ºC and centrifugation for 10 min at 12 000 rpm (Martinez et al., 2009). Protein concentrations were determined using the BioRad kit, and BSA as a standard. Protein profiles of different reserve protein fractions of barley grain, during the imbibition time, were analysed using standard SDS–polyacrylamide gels stained with Coomassie Brilliant Blue R-250.
Protease activities in barley grain
Electrophoretic detection of proteolytic activities was performed using mildly denaturing gelatine–polyacrylamide gels [0.1% (w/v) gelatine containing 0.1% (w/v) SDS, 12% (w/v) polyacrylamide] (Lantz and Ciborowski, 1994). Protein extracts (5 µg) from embryoless grains and isolated embryos were diluted 2-fold in electrophoresis buffer containing 62.5 mM TRIS-HCl pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 0.05% (w/v) bromophenol blue, and subjected to electrophoresis using a Bio-Rad Mini-Protein II Electrophoresis Cell System. After migration at 4 ºC, gels were transferred into a 2.5% (v/v) aqueous solution of Triton X-100 for 30 min at room temperature, to allow restoration of the proteases. Gels were then placed in an activation buffer (0.1 M phosphate pH 6.0 with 10 mM cysteine) for 20 h at 30 ºC. Proteolysis was stopped by transferring the gels to a staining solution of 0.3% (w/v) Coomassie Brilliant Blue R-250 in 40% (v/v) methanol and 10% (v/v) acetic acid. The gels were destained in 25% (v/v) methanol and 10% (v/v) acetic acid. Bands of proteolytic activity were visualized against the blue background of the gel.
To determine the types of protease activities, specific protease inhibitors {100 μM E-64 [l-trans-epoxysuccinyl-leucylamido-(4-guanidino)-butano]; 250 µM PMSF (phenylmethylsulphonyl fluoride), and 20 µM pepstatin-A} were added to the samples and incubated at 37 ºC for 30 min before loading onto the gel. After migration at 4 ºC, gels were treated in the same way as those without inhibitors.
To quantify the effect of the synthetic E-64 CysProt inhibitor, in vitro inhibitory assays were performed with protein extracts prepared from de-embryonated grains and isolated embryos in a buffer containing 100 mM sodium phosphate pH 6.0, 10 mM l-cysteine, 10 mM EDTA, and 0.01% (v/v) Brij35, using the Z-FR-AMC substrate (25 µM). The reactions were incubated for 1 h at 30 ºC and the emitted fluorescence was measured as described above.
Hydrolysis of stored proteins in barley grains
The capacity of the barley HvPap-1 CysProt to degrade grain storage proteins was checked by incubating 6 µg of different barley protein fractions (hordeins, albumins, and globulins) with 3 µg of the recombinant protease for 4 h at 30 ºC. The effect of barley cystatins and the HavPap-1 propeptide on its cognate HvPap-1 enzyme in the major barley reserves were determined by in vitro assays. Recombinant purified cystatins (HvCPI-1, -2, -5, -6, and -11) at 3 µM, and different concentrations (3, 5, and 10 µM) of HvPap-1pro and its derived variant HvPap-1pro* purified from E. coli cultures, were independently incubated with HvPap-1 protease for 30 min prior to the addition of the hordein storage fraction. Then, mixtures were electrophoretically separated by SDS–PAGE and stained with Coomassie Brilliant Blue R-250.
Specimen processing for microscopy
Barley embryos 24 hai were fixed in a freshly prepared solution of 4% (v/v) formaldehyde in phosphate-buffered saline (PBS) under vacuum and left overnight at 4 ºC. The samples were washed in PBS and dehydrated in a series of increasing concentrations of methanol as follows: methanol 30% (v/v) 30 min, methanol 50% (v/v) 30 min, methanol 70% (v/v) 30 min, and methanol 100% 90 min (with three changes) at 4 ºC. Then, the specimens were progressively infiltrated in LRwhite resin (Agar Scientific) in a series of mixtures of methanol:LRwhite with increasing concentrations of resin [2:1, 1:1, and 1:2 (v:v)], for 1 h each at 4 ºC, then left overnight in pure resin with 0.5% (v/v) bezoin-methyl-ether as a catalyst at 4 ºC. Polymerization in capsules was performed under UV light at –20 ºC for 2 d and at 22 ºC for 1 d. Sections of 1–2 µm were cut from the polymerized blocks in a Leica EM UC6 ultramicrotome. For structural analysis and evaluation of the fixation, the sections were stained with either 1% (w/v) toluidine blue O (Panreac) or Coomassie Brilliant Blue R-250 (diluted 1:2 in water from the solution used to stain the protein gels), observed with a Zeiss Axiophot microscope under differential interference contrast (DIC) (toluidine blue) or bright field (Coomassie), and photographed using a Leica DFC300 FX CCD camera under the Leica Application Suite 2.8.1 build 1554 acquisition software. Final images were composed with Adobe Photoshop 7.0.
Immunofluorescence detection of HvPap-1
For immunofluorescence, 1–2 µm thin sections were collected on 10-well Teflon-printed slides (Fisher Scientific Inc.) and treated with PBS for 5 min and 5% (w/v) BSA in PBS for 10 min. Then, they were incubated with 20 µl drops per well of a 1/200 solution of the rabbit-raised antibody to the HvPap-1 (HvPap-1-IgG) in PBS, for 1 h at room temperature in a humid chamber. After two washes of 15 min in PBS, an Alexa Fluor 488 anti-rabbit antibody (Molecular Probes) was applied in a 1/25 solution in 2.5% (w/v) BSA in PBS for 45 min at room temperature in a humid chamber. Subsequent to another two washes with PBS for 15 min each, the slides were mounted in a 50:50 solution of glycerol:PBS and observed with a Zeiss Axiophot microscope under blue light with the filter set ex 450–490, FT 510, em LP 520. To determine the background due to naturally autofluorescent structures, the sections were also examined under UV light (filter set ex BP 365, FT 395, em LP 397) and green light (filter set ex BP 546, FT 580, em LP 590). The structures underlying the fluorescent labelling were revealed by DIC. The images were captured using a Leica DFC300 FX CCD camera under the Leica Application Suite 2.8.1 build 1554 acquisition software. Negative controls were performed avoiding the primary antibody.
Results
A cathepsin F-like protease is involved in the germination of the barley grain
A detailed transcriptome analysis of barley grain germination using the Affymetrix Barley1 GeneChip was previously performed (Sreenivasulu et al., 2008). Based on these data, several C1A papain-like CysProts were detected to be highly expressed in both the aleurone and the embryo during grain germination. Among them, a protein (probe Contig2402_s_at) was determined to belong to the subgroup of cathepsin F-like CysProts, which had not been previously characterized in the barley grain. This protein corresponds to the HvPap-1 protein detected in barley (Martinez and Diaz, 2008). To corroborate its presence in the germinating grain, barley de-embryonated grains were subjected to treatment with 1 µM GA for different time periods (8, 24, and 48 hai). The aleurone layers were isolated and the expression pattern was analysed by qRT-PCR. As shown in Fig. 1A, the HvPap-1 gene was expressed during grain germination, and GA treatment induced a remarkable increase in its expression, which peaked at 24 hai.
Like other C1A peptidases, HvPap-1 is synthesized as an inactive precursor which comprises a signal peptide, an N-terminal propeptide of 217 amino acids, and the mature protein, with an estimated size of 40.016 kDa (Fig. 1B). The propeptide contains the non-contiguous ERFNAQ signature (Ex3Rx3Fx3Nx3Ax3Q) specific to cathepsin F-like proteases, and the consensus motif, GxN/TxFxD, common to all CysProts. Interestingly, a QVVGG sequence, which is the conserved inhibitory site of cystatins (Martinez and Diaz, 2008), responsible for their inhibitory capacity against C1A CysProts, appears at the N-terminal part of the propeptide. In the mature protein, the active site residues C166, H308, and N335, crucial in the activity of these peptidases, the GCNGG like-motif, common to all CysProts, and several cysteine residues, presumably involved in the formation of disulphide bridges to maintain the three-dimensional structure of the protein, were identified. In addition, the analysis of the HvPap-1 sequence predicted the presence of a potential N-glycosylation site NYS in the mature protein (Fig. 1B).
To show how the sequence determined the three-dimensional structure, the known crystal structure of the human cathepsin-F (Wang et al., 1998) was used as a molecular model to establish the predicted structure of the barley HvPap-1 (Fig. 1C). Globally, both models were similar, although several changes in the location and size of secondary structures such as some α-helices and β-sheets were detected. These differences putatively imply different functional specificities for the substrates they could degrade.
Evolutionary insights into cathepsin F-like proteases in angiosperms
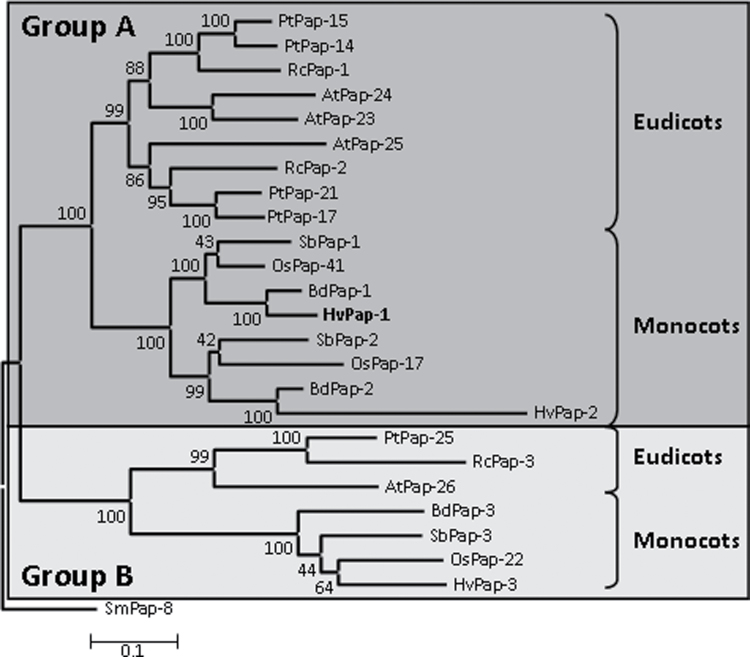
An evolutionary analysis of papain-like CysProts, including cathepsin F-like proteins, from algae to land plants, was previously reported (Martinez and Diaz, 2008). To obtain more information on the evolutionary relationships of cathepsin F-like proteins in angiosperms, three monocot (B. distachyon, O. sativa, and S. bicolor) and three eudicot species (A. thaliana, P. trichocarpa, and R. communis) were selected. The genomes of these six species have been completely sequenced and drafts of these sequences are available on the internet. Therefore, searches by BLAST were made to obtain the cathepsin F-like proteases from them (Table 1). Sequences that presented the conserved ERFNAQ motif were selected. The sequences of cathepsin F-like proteases from H. vulgare were also retrieved (Martinez and Diaz, 2008). To obtain evolutionary information on this family in angiosperms, the selected proteins were aligned by MUSCLE (Supplementary Fig. S1), and a phylogenetic tree was constructed by the Neighbor–Joining method (Fig. 2). The angiosperm cathepsin F-like sequences were grouped into two different clades supported by bootstrap values of 100%. Both clades included proteins from all eudicot and monocot species. Group A was formed by two subgroups; one of them contained eudicot sequences and the other monocot sequences. The composition of the subgroups suggests a duplication event in the ancestor both of the eudicot and of the monocot species. The sequences in group B were also separated into eudicot and monocot clades. Only one sequence per species was detected, suggesting orthology for all group B proteins. The barley cathepsin F-like protein HvPap-1 belonged to a clade within group A together with BdPap-1, OsPap-41, and SbPap-1, putative orthologues in cereal species.
Table 1.
Inhibition constant (Ki ) values of the barley cystatins and the two propeptides derived from the HvPap-1 protease, against the HvPap-1 barley peptidase expressed as recombinant proteins in E. coli cultures
| Inhibitors | Ki (M) |
| Cystatins | |
| HvCPI-1 | 1.5×10-7 |
| HvCPI-2 | 4.1×10-8 |
| HvCPI-3 | NI |
| HvCPI-4 | 2.8×10-7 |
| HvCPI-5 | 3.5×10-7 |
| HvCPI-6 | 1.3×10-9 |
| HvCPI-7 | NI |
| HvCPI-8 | 6.4×10-7 |
| HvCPI-9 | 1.8×10-8 |
| HvCPI-10 | NI |
| HvCPI-11 | 2.5×10-8 |
| HvCPI-12 | NI |
| HvCPI-13 | 7.3×10-7 |
| Propeptides | |
| HvPap-1pro | 2.3×10-8 |
| HvPap-1pro* | 1.5×10-8 |
No inhibitory activity (NI) was observed at 5 mM concentration of the cystatin.
Fig. 2.
Phylogram of the cathepsin F-like CysProts from several monocot and eudicot species. The amino acid sequences were aligned by MUSCLE and analysed with the Neighbor–Joining method. Bootstrap values are indicated. A cathepsin F-like protein from Selaginella moellendorffii (SmPap-8) was used to root the tree. At, Arabidopsis thaliana; Pt, Populus trichocarpa; Rc, Ricinus communis; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Os, Oryza sativa; Sb, Sorghum bicolor.
Recombinant active HvPap-1 protease is correctly processed
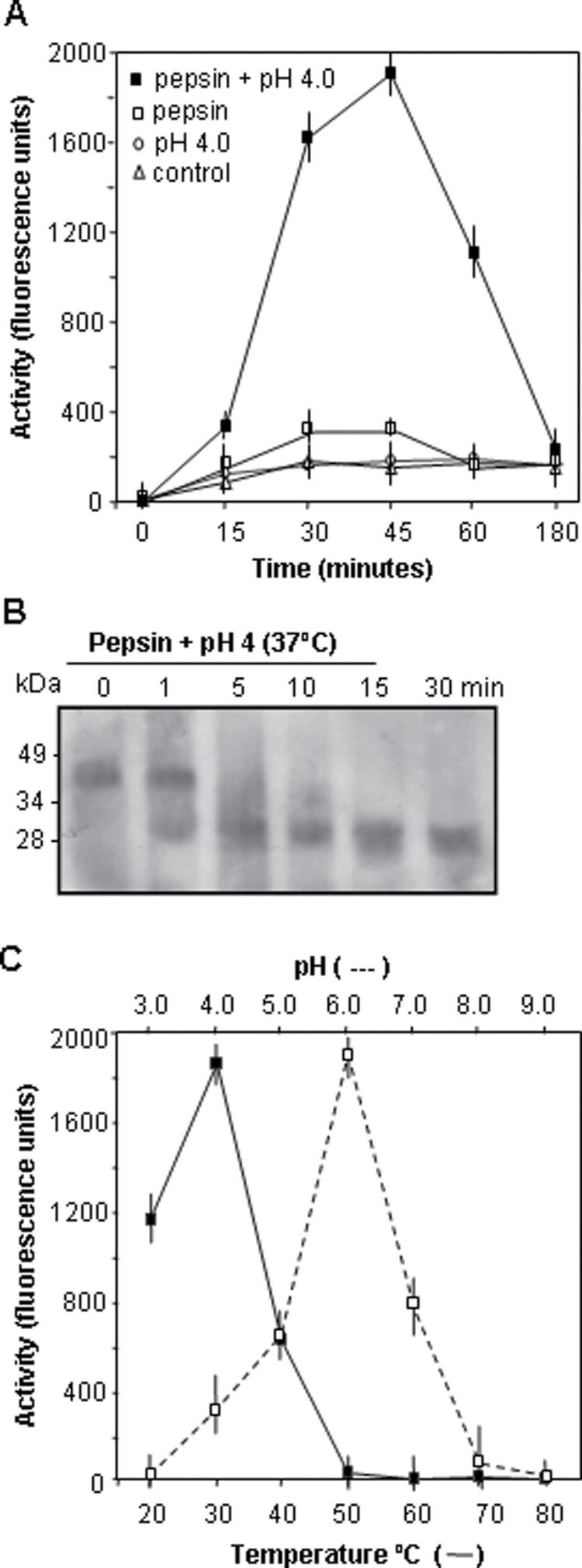
To analyse further the protease properties of HvPap-1 protein, it was expressed as a fusion protein with a histidine tail in E. coli cultures. The recombinant protein was purified to homogeneity by denaturing Ni-NTA affinity chromatography, and refolded by rapid dilution into a buffer at pH 8.0, yielding folded HvPap-1. Since some related cysteine endoproteases self-activate in vitro to their mature forms in an acidic environment while other enzymes need an assistant protease to be activated, an analysis was carried out to determine whether recombinant HvPap-1 could be activated to its mature form under analogous conditions. The recombinant HvPap-1 was therefore incubated in a pH 4.0 buffer with and without pepsin. At the same time, several fluorescence substrates susceptible to degradation by CysProts were assayed to determine the most suitable for HvPap-1 studies. Proteolytic activity was only observed with a combination of low pH and pepsin (Fig. 3A), and its maximum levels were reached when the Z-FR-AMC substrate was assayed (Supplementary Fig. S2 at JXB online). The extent of activation as a function of time was analysed by SDS–PAGE. As shown in Fig. 3B, under these conditions, the HvPap-1 precursor of ~40 kDa was almost completely processed to its mature form (~26 kDa) within the first 5 min. Likewise, the activity was higher when reactions were incubated at pH 6.0 and a temperature of 30 ºC (Fig. 3C).
Fig. 3.
Analysis of the biochemical parameters of the recombinant barley HvPap-1 protease. (A) Time-dependent activation of the barley procathepsin F-like HvPap-1 peptidase at 37 ºC in 100 mM sodium acetate buffer pH 4.0 with 0.6 mg ml–1 pepsin (filled squares), in 100 mM sodium acetate buffer pH 4.0 (open circles), in 100 mM TRIS-HCl buffer pH 8.0 with 0.6 mg ml–1 pepsin (open squares), and in 100 mM TRIS-HCl buffer pH 8.0, considered as control (open triangles). Values, expressed as units of fluorescence emitted at 465 nm, are means ±SE of triplicate measurements. (B) Western blot analysis of the recombinant proprotein HvPap-1 pepsin-mediated processing at pH 4.0, at different incubation times (in minutes). (C) pH and temperature profiles for the recombinant barley HvPap-1 protease activity.
Recombinant HvPap-1 propeptide and cystatins are able to inhibit HvPap-1 protease activity
To test the inhibitory capacity of the HvPap-1 propeptide against its cognate HvPap-1 enzyme, two different constructs were prepared and expressed as recombinant proteins. HvPap-1pro, which spanned the entire HvPap-1 propeptide (A25–G141) and HvPap-1pro* which lacked the first 19 amino acid residues (D44–G141) to exclude the QVVGG motif, described above as the conserved inhibitory site of cystatins, were expressed in E. coli as fusion proteins and purified to homogeneity by Ni2+ affinity chromatography (Supplementary Fig. S3 at JXB online). Inhibitory assays were done against HvPap-1 peptidase using the optimal conditions described above. Kinetic analyses revealed that both HvPap-1 propeptides, HvPap-1pro and HvPap-1pro*, exhibited a competitive tight binding inhibition against HvPap-1. Likewise, K i values were similar for both propeptide constructs (Table 1).
Additionally, we also tested the capability of the barley cystatins to block the HvPap-1 protease activity. Inhibitory in vitro assays were done using the conditions previously established. Kinetic analyses revealed that barley cystatins also exhibited a competitive inhibition against HvPap-1. The inhibition constant values (K i) were determined, and showed different specificities (Table 1). Nine of the 13 barley cystatins were good inhibitors of HvPap-1, cystatin HvCPI-6 being the strongest inhibitor with a K i value of 3.9×10–9 M. Barley cystatins HvCPI-3, -7, -10, and -12 were inactive towards HvPap-1 protease under the tested conditions.
HvPap-1 is ubiquitously expressed
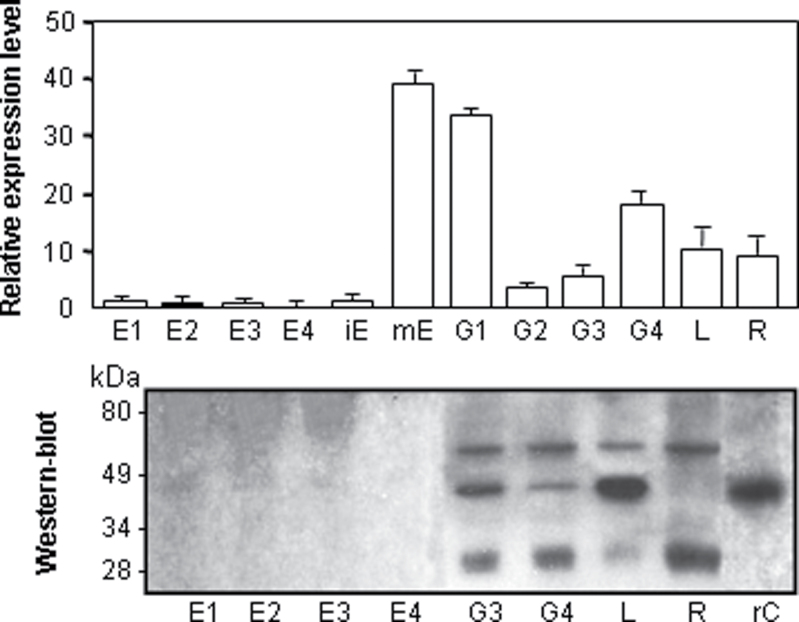
To study the expression pattern of the HvPap-1 gene and its HvPap-1 protein, qRT-PCR normalized to barley Actin2 mRNA levels and western blot analyses using a specific HvPap-1 antibody were carried out in the major barley tissues. RNA and protein samples were prepared from developing endosperm at 10, 14, 18, and 22 daf, from immature (18 daf), mature, and germinating embryo at 8, 16, 24, and 48 hai, and from 7-day-old leaves and roots (Fig. 4). The HvPap-1 gene was mainly expressed in the mature and germinating embryo and in the vegetative tissues such as the leaf and root. Meanwhile, its expression was scarcely detected in the endosperm and in the immature embryo. These results were confirmed by western blot assays where a direct correlation between the transcript and the protein accumulation patterns could be appreciated. Interestingly, the protein profile showed three bands of different sizes. The smallest and the middle size bands of ~26 kDa and 40 kDa corresponded to the mature processed protein and the immature protease containing the propeptide, respectively. The higher size band could be due to post-translational modifications which did not take place when the protease HvPap-1 was purified as a recombinant protein in E. coli cultures (lane rC in Fig. 4).
Fig. 4.
Analysis of the mRNA and protein expression of the barley HvPap-1 gene, assayed by real-time quantitative PCR and western blot analysis, respectively. Expression of the HvPap-1 gene in different barley tissues. RNAs and proteins were extracted from developing endosperm at 10 (E1), 14 (E2), 18 (E3), and 22 (E4) daf, from immature embryos (iE), mature embryos (mE), and germinating embryos collected at 8 (G1), 16 (G2), 24 (G3), and 48 (G4) hai, and from 7-day-old leaves (L) and roots (R). Recombinant proprotein HvPap-1 was purified from E. coli cultures (rC). Values of transcripts, expressed as the relative mRNA content of the cathepsin F-like HvPap-1, were standardized to the barley Actin2 mRNA level. The approximate location of molecular marker bands for western blots is shown. The polyclonal antibody HvPap-1-IgG was prepared against a specific peptide, indicated in Fig. 1B.
HvPap-1 cathepsin F-like protease is N-glycosylated
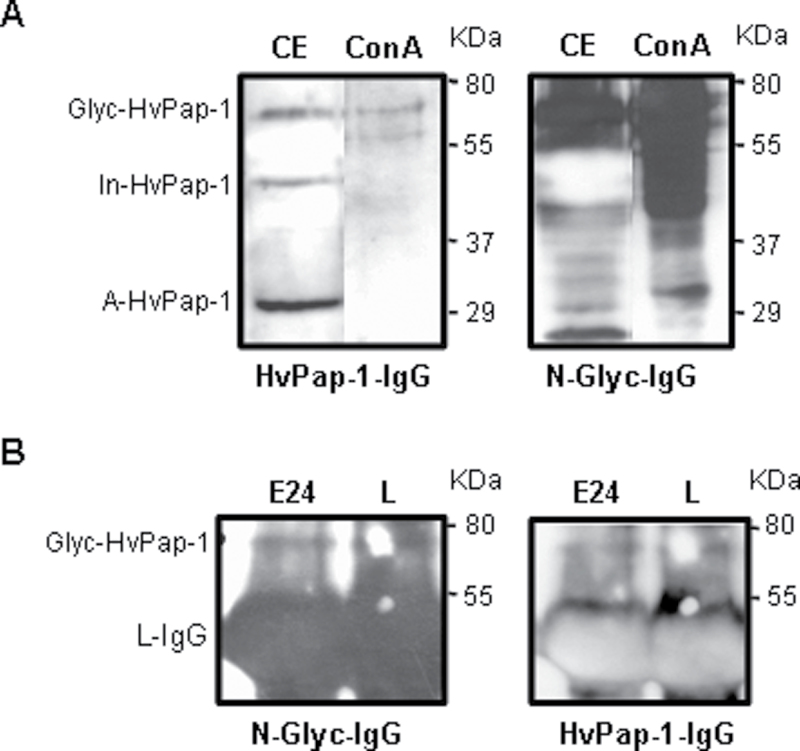
The absence of the highest size protein band observed in the western blot assays when the HvPap-1 protease was purified as a recombinant protein in E. coli cultures suggested that a post-translational glycosylation of the protein occurred. To analyse the putative glycosylation state of HvPap-1 protease, two approaches were used. First, total glycoproteins from barley leaves were isolated using Con A affinity column chromatography. Immunoblotting profiles of crude leaf proteins and glycoprotein preparations were compared using the specific HvPap-1-IgG antibody and an anti-N-glycosylation N-Glyc-IgG antibody. As shown in Fig. 5A, the putative glycosylated form of HvPap-1 observed in crude extracts was coincident with a similar size band detected by the antibody HvPap-1-IgG in the leaf glycoprotein sample. As expected, a complex pattern of proteins appeared when membranes were incubated with anti-N-glycosylation antibody (N-Glyc-IgG). However, these results showed the high specificity of the HvPap-1-IgG antibody able to recognize the putative glycosylated HvPap-1 within a complex pool of glycoproteins. Secondly, to assess the existence of the glycosylated protease form, the native HvPap-1 protein was isolated by immunoprecipitation from crude barley extracts (embryo and leaf) using the specific antibody HvPap-1-IgG. The immunoprecipitates were analysed by western blot assays with both antibodies, HvPap-1-IgG and N-Glyco-IgG. Interestingly, the immunoblotting revealed the presence of the same band pattern in germinating embryo and leaf samples independently of the antibody used (Fig. 5B), indicating that the highest size band corresponded to the glycosylated form of the HvPap-1 protein. The band which was smaller in size, but of much greater intensity, corresponded to the large subunit of the HvPap-1-IgG. These results indicated the presence of an N-glycosylated form of the HvPap-1 peptidase which is supported by the N-glycosylation site identified in its amino acid sequence (Fig. 1B).
Fig. 5.
Analysis of the glycosylation state of HvPap-1 protease. (A) Western blot analysis of crude protein extracts from leaves (CE) and glycoproteins purified from barley leaves on a concanavalin A column (Con A). Membranes were incubated with anti-HvPap-1 (HvPap-1-IgG) and anti-N-glycosylation (N-Glyc-IgG) antibodies. (B) Protein extracts from 24 h germinating embryos (E24) and 7-day-old barley leaves (L) were immunoprecipitated with the HvPap-1-IgG antibody and analysed by western blot. Membranes were first incubated with the HvPap-1-IgG antibody, then stripped, and subsequently the N-Glyc-IgG antibody was used. Glycosylated (Glyc-HvPap-1), inactive (In-HvPap-1), and active (A-HvPap-1) forms of the HvPap-1 protease are indicated. L, large subunit IgG immunoglubulin.
CysProt activity is present in the barley grain during germination
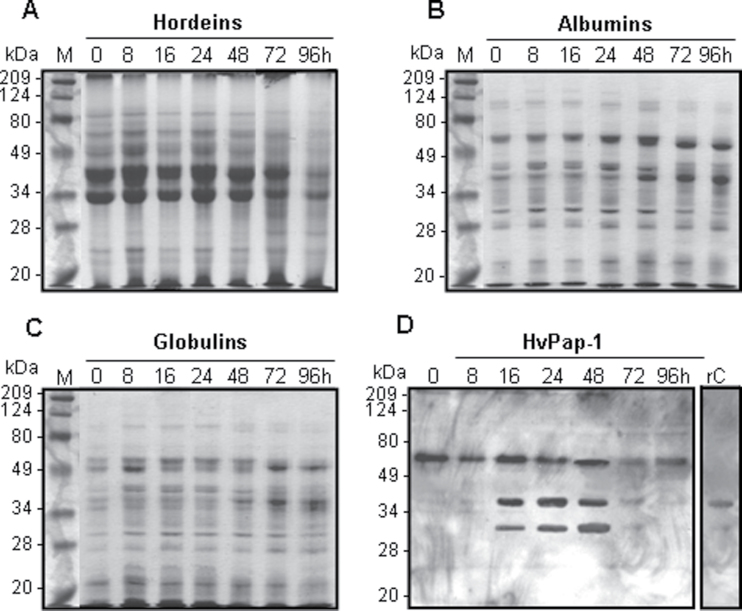
Due to HvPap-1 protein being detected in most tissues of the barley grain (Fig. 4), the protein pattern and the proteolytic activities were further analysed in two grain fractions (de-embryonated grain and embryo) during germination. Based on hordeins being the most abundant proteins accumulated in the mature barley grain together with albumins and globulins, enriched fractions of the three types of reserve proteins were prepared at different germination times. Protein fractions were electrophoretically separated and, as is shown in Fig. 6A–C, the degradation pattern profile of hordeins, albumins, and globulins during the imbibition process was detected at 24–48 hai and was greatly increased until 96 hai. The reduction of protein bands of larger size was parallel to an increase in smaller size bands between 48 and 96 hai. Their temporal pattern of degradation correlated with the production of the active HvPap-1 CysProt form detected from 16 hai by western blot assays (Fig. 6D). In addition, total crude proteins were extracted from embryos at different times after water imbibition and the protein degradation products were also observed after 24 h of water imbibition (Supplementary Fig. S4A at JXB online).
Fig. 6.
Protein pattern from de-embryonated barley grain during germination. (A–C) Protein accumulation pattern of reserve proteins (hordeins, albumins, and globulins) from barley de-embryonated grain at different germination times (0–96 h) stained with Coomassie Brilliant Blue R-250. (D) Expression of HvPap-1 protein of de-embryonated barley grain at different germination times (0–96 hai) using the HvPap-1-IgG antibody.
To study the proteolytic activities involved in grain protein degradation, the activity of the main protease groups (cysteine, serine, and aspartic proteases) was analysed in the same two grain fractions. Protease activities were assayed using mild denaturing gelatine–PAGE. After electrophoretic protein separation and subsequent renaturation, active proteases hydrolysed gelatine, showing clear bands of lysis against dark Coomassie Blue-stained gelatine in the gel. As shown in Supplementary Fig. S5 at JXB online, the band profile showed the major protease activity between 48 and 72 hai and drastically decreased at 96 hai. Curiously, an intense activity band was also detected before starting the water imbibition (0 hai), indicating the accumulation of proteases in the mature grain. When protein extracts were pre-incubated with the CysProt inhibitor E-64, the intensity of the hydrolytic bands was reduced and some bands even disappeared. The inhibition was greater after pre-incubation in the presence of the PMSF protease inhibitor which is considered as specific for serine proteases although it can also partially inhibit CysProts (Dunn, 1989). In contrast, the specific aspartic protease inhibitor pepstatin did not inhibit the proteolytic process (Supplementary Fig. S5).
Assays carried out with protein extracts prepared from isolated embryo fraction showed a hydrolytic activity-associated band at 48–96 hai which was slightly reduced in the presence of E-64 (Supplementary Fig. S4B, C at JXB online). The quantification of proteolytic activity by an inhibitory in vitro assays with or without the E-64 inhibitor corroborated the cysteine-type protease activity already detected at 24 hai, with a peak at 72 hai for embryo extracts and at 96 hai or even later for the de-embryonated grain fraction (Supplementary Fig. S6).
HvPap-1 is involved in the degradation of storage proteins in the barley grain
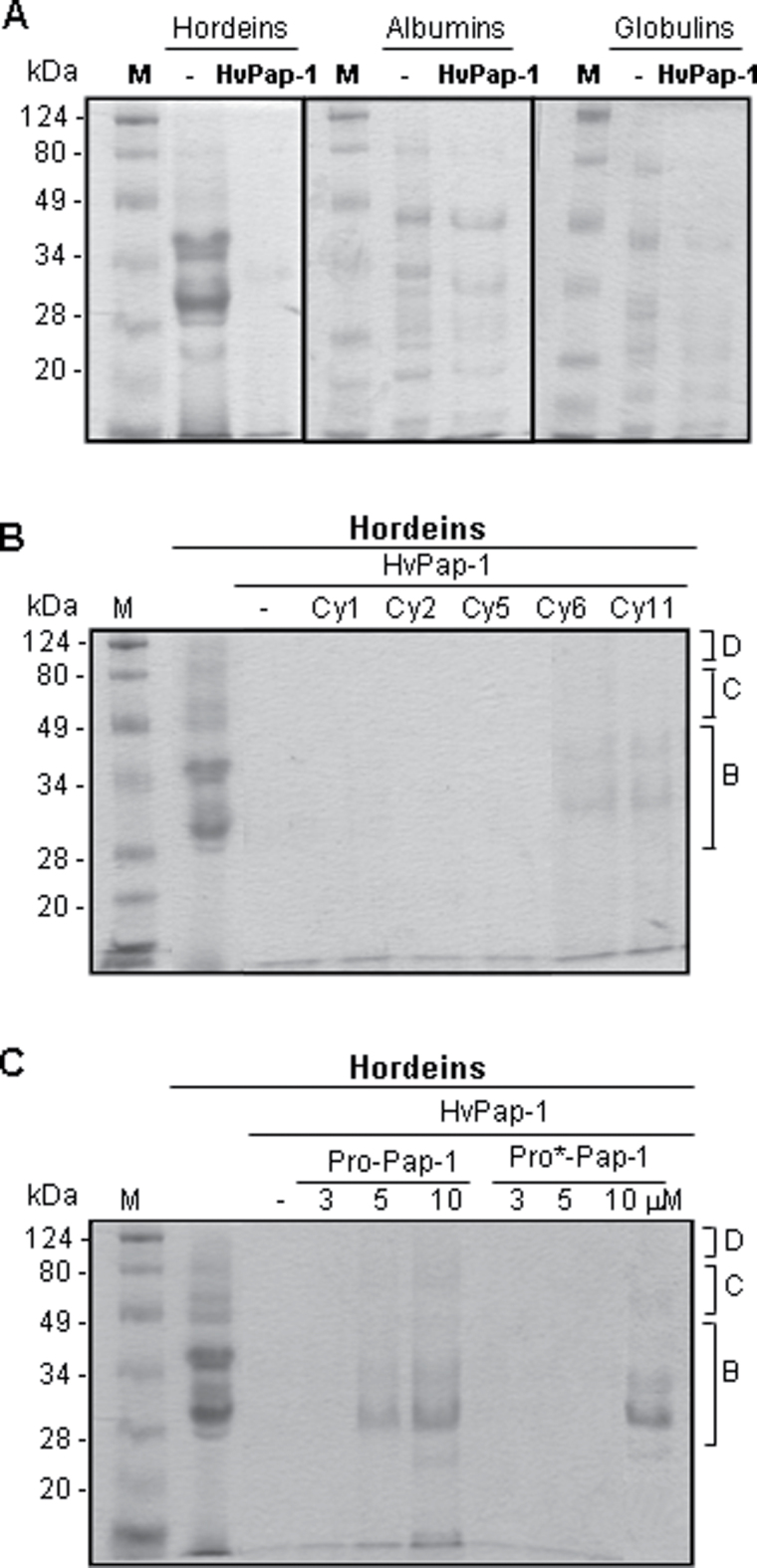
The functional relevance of CysProt activity detected in the two fractions of barley grain was investigated by analysing the implication of HvPap-1 in the mobilization of different storage proteins during grain germination. When different reserve protein types (hordeins, albumins, and globulins) were extracted and incubated with the recombinant HvPap-1 CysProt (Fig. 7A), the barley cathepsin F-like peptidase totally hydrolysed all electrophoretic bands corresponding to B, C, and D hordeins, but only a partial degradation of albumin and globulin bands was detected at the concentration tested. Additionally, whether the combination of HvPap-1 with barley cystatins and with its own propeptide modified the breakdown of protein reserves accumulated in the endosperm was explored. The results indicated that the capability of HvPap-1 CysProt to degrade hordeins was not altered by the addition of the barley recombinant cystatins HvCPI-1, HvCPI-2, HvCPI-5, HvCPI-6, and HvCPI-11 at 3 µM concentration. Only some weak bands appeared in the presence of cystatin HvCPI-6 and HvCPI-11 (Fig. 7B). When propeptide HvPap-1pro or its variant HvPap-1pro* were incubated with the HvPap-1 protease prior to the addition of the hordein fraction, the proteolytic activity was not inhibited at low propeptide concentrations (3 µM). However, higher concentrations of propeptides showed a recovery of the hordein band profiles (Fig. 7C).
Fig. 7.
Effects of barley HvPap-1 CysProt, barley cystatins, and HvPap-1 propeptide on grain reserve mobilization. (A) Effects of barley HvPap-1 protease on the degradation of grain storage proteins. Proteins (hordeins, albumins, and globulins) extracted from barley (cv. Bomi) dry grains were incubated with 3 μg of the recombinant protease HvPap-1. (B) Effect of 3 μM barley cystatins HvCPI-1, -2, -5, -6, and -11 (Cy1, Cy2, Cy5, Cy6, and Cy11) on the proteolytic activity of HvPap-1 to degrade hordeins. (C) Effect of different concentrations of the HvPap-1 propeptides (HvPap-1pro and HvPap-1pro*) on the proteolytic activity of HvPap-1 to degrade hordeins. SDS–polyacrylamide gels were stained with Coomassie Brilliant Blue R-250. Molecular masses of molecular markers (M) are shown in kDa. The location of B, C, and D hordeins are indicated.
HvPap-1 is located in cytoplasmic refringent bodies
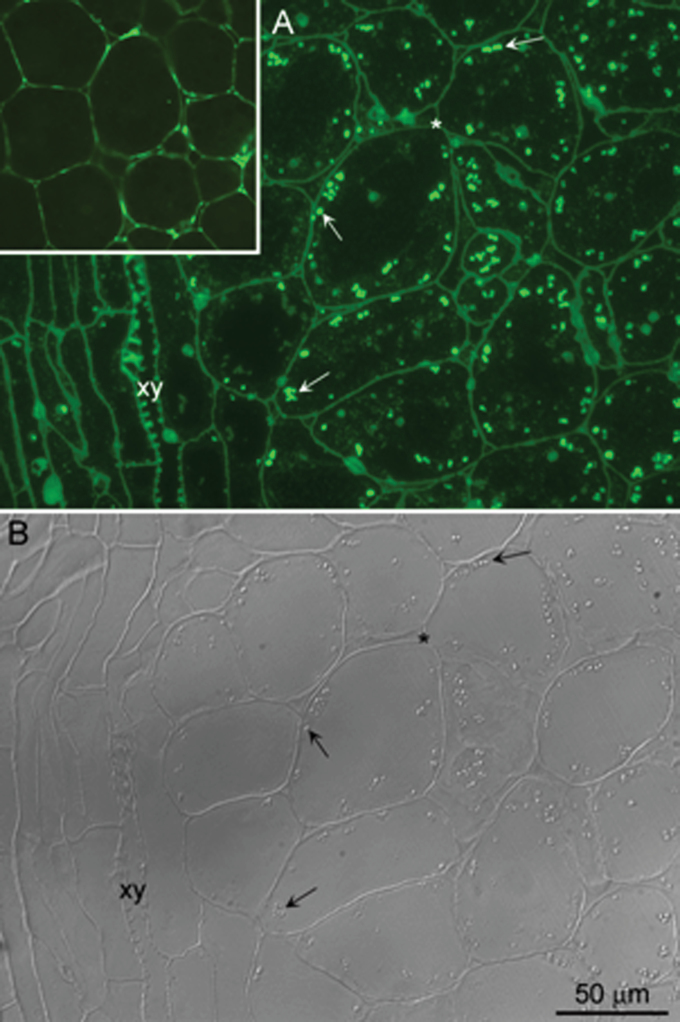
To determine the subcellular location of the HvPap-1 protease in the grain, immunodetection assays were performed in germinating embryos as the most important living grain tissue. A structural check of the specimens to be used for immunofluorescence showed a good fixation of the embryo, with polygonal-shaped cells containing a large nucleus and a cytoplasm rich in vacuoles of different sizes and round refringent bodies detected by DIC (Supplementary Fig. S7A at JXB online). Coomassie staining on parallel sections identified numerous cytoplasmic protein bodies with a similar size to that of the refringent structures revealed by the DIC technique (Supplementary Fig. S7B). Immunofluoresence experiments with a specific antibody against the C-terminal peptide of HvPap-1, HvPap-1-IgG antibody previously used for western blot and glycosylation assays, showed abundant bright foci dispersed in the cytoplasm as ring-shaped structures (arrows in Fig. 8A; Supplementary Fig. S8B). The corresponding image of the cell’s structure under DIC revealed that the fluorescence was located in the refringent bodies (arrows in Fig. 8B). Some unspecific labelling was also found in the naturally autofluorescent cell walls, in lignin of xylem vessels, and in deposits inside the vacuoles (Fig 8A; Supplementary Fig. S8B). Unspecific labelling due to cell wall components was also seen in the negative controls without the primary antibody (insert in Fig. 8A). These structures showed a fluorescent signal when excited with UV, blue, and green light (Supplementary Fig. S8A, C). The specific signal detecting the HvPap-1 protease was only observed under blue light. Negative controls without the primary antibody did not show any labelling (data not shown).
Fig. 8.
Immunodetection of HvPap-1 protease in barley embryos after 24 h of water imbibition. (A) Immunofluorescence localization of HvPap-1 on 2 μm sections of formaldehyde-fixed and LR white-embedded barley embryos 24 hai. Numerous bright foci of different sizes (arrows) are observed within the cytoplasm, excluding the vascular bundle (xy). (B) The fluorescent foci correspond to refringent structures (arrows) revealed by differential interference contrast (DIC). The cell walls (asterisk), clearly identified by DIC, also show some unspecific autofluorescence as seen in the negative control (inset in A).
Discussion
Barley grain has been considered as a monocot model in germination research and, like other cereals, stores starch, proteins, and lipids in the endosperm that are used during the germination process until photosynthesis is fully established. Barley germination starts with water uptake by the quiescent dry seed and ends with protrusion of the radicle tip through the grain coat 16–24 h after imbibition. During this phase, the embryo produces phytohormones, mainly GAs, which induce expression and/or de novo synthesis of hydrolytic enzymes in aleurone layers. In a second phase, the enzymes are secreted and break down the stored reserves of the endosperm, releasing nutrients that support embryo growth until the onset of photosynthesis (Bove et al., 2002; An and Lin, 2011). Most studies have been focused at the transcriptional level involving a complex network of regulatory programmes (An and Lin, 2011). In contrast, remarkably little is known about post-germination processes where proteases and amylases involved in hydrolysing and mobilizing storage compounds are also strictly regulated. The contribution of C1A CysProts of barley in post-germination events has been shown, but to date only members of the cathepsin L-, H-, and B-like groups involved in this process have been characterized (Chen and An, 2006). The HvPap-1 protein, described in this study, belongs to the cathepsin F-like C1A CysProt group (Martinez and Diaz, 2008). It first appeared as a non-characterized enzyme (probe Contig2402_s_at) among protease genes during grain germination identified in the transcriptome analysis of the barley grain (Sreenivasulu et al., 2008). Here it is shown that the HvPap-1 gene/protein is expressed mainly, although not exclusively, in germinating grain tissues. In particular, it has been detected in embryo and aleurone layers where it is induced after incubation in the presence of GAs. Moreover, the immunodetection assays of germinating embryo have localized cathepsin F-like HvPap-1 in small, refringent structures of the size of those stained with Coomassie, probably protein bodies. This subcelullar location is determined by the N-terminal signal peptide of the HvPap-1 protein that directs the nascent protein to the ER, where a glycosylation event takes place in a unique site found in its sequence. The lack of the KDEL retention signal suggests a further movement through the secretory pathway, presumably via Golgi vesicles, and loading into embryo protein bodies upon germination. Maturation of ER-synthesized N-linked glycosylated proteins and de novo O-glycosylation within the Golgi apparatus fulfil major roles in protein sorting through the endomembrane system by providing tags for efficient subcellular fate (Schoberer et al., 2009). Thus, the subcellular traffic experienced by HvPap-1 protease to reach its final location and its glycosylated form are two closely related physiological events.
Mature barley grains accumulate hordeins, albumins, and globulins in the protein bodies, as the main reserve proteins which were the appropriate targets of proteases post-germination to support the first steps of seedling growth. However, little is known about hydrolases synthesized by the aleurone layer, with the exception of starch-degrading enzymes which have been studied in detail due to their importance for the malting industry. The prevalence of serine and cysteine peptidase activities detected in the germinating embryo and aleurone layer in the present experiments using zymograms and fluorescent substrates implicated these enzymes in the mobilization of reserves. A recent study of intracellular and secreted proteomes analysed in barley aleurone layer identified one serine-carboxy-peptidase and nine extra- and three intracellular C1A CysProts (Finnie et al., 2011), but none of them corresponds to the cathepsin F-like HvPap-1. However, the same authors highlighted the extreme difficulties in detecting secreted proteins from aleurone during germination since most of them are produced in much smaller quantities in comparison with the abundant α-amylases. Nevertheless, the temporal coincidence of depletion of grain protein reserves with the expression pattern of HvPap-1 peptidase and, in particular, with its temporal protein processing to generate mature peptidase forms, demonstrates that HvPap-1 plays an active role in the post-germination process.
Plant C1A CysProts are synthesized as inactive or weakly active precursors to prevent inappropriate proteolysis and to become active when they need to be processed. Therefore, the prosequences act as important modulators of the protease activity to guarantee that the mature enzyme is formed in the right place and/or at the right time (Demidyuk et al., 2010). The protease precursor activation is a complex process involving the appropriate pH, other protease activities, inhibitors, and some compounds strongly dependent on the cellular or extracellular environment. A key point for the further analysis of protein processing and activity of a peptidase is to be able to purify it. To date, other cathepsin F-like proteins from the human liver fluke Opisthorchis viverrini and the olive flounder Paralychthys olivaceous have been purified (Wang et al., 1998; Ahn et al., 2009; Pinlaor et al., 2009). For the first time, a plant cathepsin F-like protein as well as its propeptide has been purified in E. coli. Recombinant proteins have allowed assays not only on protein processing but also on peptidase interactions with their putative inhibitory proteins (cystatins and propeptides). In fact, in vitro inhibitory assays have been performed using the whole family of barley cystatins (Martinez et al., 2009) and its own inhibitory propeptide, both purified as recombinant proteins. Moreover, two different constructs of the HvPap-1 propeptide were prepared, which differed in the presence of a QVVAG motif that resembles the reactive inhibitory site of cystatins. However, no differences in the inhibitory properties between the two variants were observed, indicating that the cystatin motif was not responsible for the inhibition. As expected, most cystatins were able to inhibit the activity of the mature HvPap-1 protease, suggesting a complex control of peptidase activity mediated by their inhibitors.
Focusing on the interaction of proteases and inhibitors in the mobilization of storage proteins, HvPap-1 degraded the most important storage proteins in barley, the hordeins, more efficiently than albumins or globulins. It has been suggested that C1A CysProts and cystatins form inactive complexes that are dissociated when they interact with the storage protein bodies (Kiyosaki et al., 2007). However, in contrast to the inhibition of cathepsin L-like activity by barley cystatins, and its in vitro capacity to inhibit HvPap-1, higher concentrations of all cystatins tested are needed to reverse the degradation of hordeins by HvPap-1 peptidase. Likewise, only high concentrations of HvPap-1 propeptide block the proteolytic action of its cognate HvPap-1 enzyme. These results reinforced the importance of further analysis of the cystatin–CysProt interaction in the regulation of proteases, and open up the possibility of an alternative regulatory mechanism involving propeptides released from them. Taking all these results together, it can be concluded, first, that barley HvPap-1 cathepsin F-like peptidase participates as an active component together with other cysteine and serine proteases in the mobilization of grain stored proteins, mainly hordeins, during the last phase of grain germination. The second, and perhaps the most important, conclusion is that HvPap-1 cathepsin F-like activity in the barley grain is strictly regulated by its subcellular location, the GA content in the grain, endogenous inhibitors such as specific cystatins, and its own propeptide.
HvPap-1 was also expressed in vegetative tissues such as leaves and roots, indicating its participation in additional physiological roles. Experiments to overexpress and silence the HvPap-1 gene by generating transgenic barley plants as well as the characterization of the other two barley cathepsin F-like proteins, belonging to the same phylogenetic group (HvPap-2) or not (HvPap-3), will bring new insights into the function of cathepsin F-like peptidases.
Supplementary Material
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Comparison of the amino acid sequences of the cathepsin-F-like cysteine proteases used in the phylogenetic tree.
Figure S2. Enzymatic activity of recombinant HvPap-1 determined using the fluorogenic substrates Z-FR-AMC, Z-RR-AMC, and Bz-FVR-AMC, susceptible to degradation by cathepsin L- B-, and H-like peptidases, respectively.
Figure S3. Purification of recombinant propeptides derived from the HvPap-1 protease.
Figure S4. Protein expression pattern and protease activity of embryos during germination.
Figure S5. Zymograms of the gelatinolytic activity of protein extracts from de-embryonated barley grains at different germination times (0–96 hai).
Figure S6. In vitro quantification of CysProt activity in extracts from de-embryonated grain and embryo fractions of barley grain at different germination times.
Figure S7. Histochemical characterization of barley grain 24 hai.
Figure S8. Autofluorescence control on sections from barley embryos 24 hai immunolabelled with the antibody to the protease HvPap-1 and revealed with Alexa Fluor 488.
Acknowledgements
We thank M. Mercedes Lucas and Susana Fajardo (CCMA-CSIC, Madrid) for the facilities provided in the preparation of the specimens for microscopy. We also greatly appreciated the anti-N-glycosylation antibody (N-Glyc-IgG) donated by Dr Araceli Diaz-Perales (CBGP-UPM-INIA, Madrid, Spain). Financial support from the Ministerio de Economia y Competitividad (project AGL2011-23650) is gratefully acknowledged.
References
- Ahn SJ, Kim NY, Seo JS, Je JE, Sung JH, Lee SH, Kim MS, Kim JK, Chung JK, Lee HH. Molecular cloning, mRNA expression and enzymatic characterization of cathepsin F from olive flounder (Paralichthys olivaceus) Comparative Biochemistry and Physiology. Part B Biochemistry and Molecular Biology. 2009;154:211–220. doi: 10.1016/j.cbpb.2009.06.005. [DOI] [PubMed] [Google Scholar]
- An YQ, Lin L. Transcriptional regulatory programs underlying barley germination and regulatory functions of gibberellin and abscisic acid. BMC Plant Biology. 2011;11:105. doi: 10.1186/1471-2229-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchabane M, Schluter U, Vorster J, Goulet MC, Michaud D. Plant cystatins. Biochimie. 2010;92,:1657–1666. doi: 10.1016/j.biochi.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. Heterologous expression, purification, refolding, and structural–functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chemistry and Biology. 2006;13:637–647. doi: 10.1016/j.chembiol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Boonmee A, Srisomsap C, Karnchanatat A, Sangvanich P. Biologically active proteins from Curcuma comosa Roxb. rhizomes. Journal of Medicinal Plant Research. 2011;5:5208–5215. [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nature Protocols . 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Bove J, Jullien M, Grappin P. Functional genomics in the study of seed germination. Genome Biology. 2002;3,reviews 1002 doi: 10.1186/gb-2001-3-1-reviews1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, An Y-QC. Transcriptional responses to gibberellin and abscisic acid in barley aleurone. Journal of Integrative Plant Biology . 2006;48:591–612. [Google Scholar]
- Demidyuk IV, Shubin AV, Gasanov EV, Kostrov SV. Propeptides as modulators of functional activity of proteases. BioMol Concepts . 2010;1:305–322. doi: 10.1515/bmc.2010.025. [DOI] [PubMed] [Google Scholar]
- Dunn BM. Determination of protease mechanism. Oxford: Oxford University Press; 1989. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban-Garcia B, Garrido-Cardenas JA, Alonso DL, Garcia-Maroto F. A distinct subfamily of papain-like cysteine proteinases regulated by senescence and stresses in Glycine max . Journal of Plant Physiology . 2010;167:1101–1108. doi: 10.1016/j.jplph.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Finnie C, Andersen B, Shahpiri A, Svensson B. Proteomes of the barley aleurone layer: a model system for plant signalling and protein secretion. Proteomics. 2011;11:1595–1605. doi: 10.1002/pmic.201000656. [DOI] [PubMed] [Google Scholar]
- Fischer J, Becker C, Hillmer S, Horstmann C, Neubohn B, Schlereth A, Senyuk V, Shutov A, Muntz K. The families of papain- and legumain-like cysteine proteinases from embryonic axes and cotyledons of Vicia seeds: developmental patterns, intracellular localization and functions in globulin proteolysis. Plant Molecular Biology. 2000;43:83–101. doi: 10.1023/a:1006456615373. [DOI] [PubMed] [Google Scholar]
- Grudkowska M, Zagdanska B. Multifunctional role of plant cysteine proteinases. Acta Biochimica Polonica. 2004;51:609–624. [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. The Plant Journal . 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Holwerda BC, Rogers JC. Purification and characterization of aleurain: a plant thiol protease functionally homologous to mammalian cathepsin H. Plant Physiology . 1992;99:848–855. doi: 10.1104/pp.99.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis IG, Drenth J, Baker EN. Thiol proteases. Comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence information for cathepsins B and H, and stem bromelain. Journal of Molecular Biology . 1985;182:317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- Karrer KM, Peiffer SL, DiTomas ME. Two distinct gene subfamilies within the family of cysteine protease genes. Proceedings of the National Academy of Sciences, USA. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Van der Hoorn RA. A model of the C14–EPIC complex indicates hotspots for a protease-inhibitor arms race in the oomycete–potato interaction. Plant Signaling and Behaviour. 2011;6:109–112. doi: 10.4161/psb.6.1.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosaki T, Matsumoto I, Asakura T, Funaki J, Kuroda M, Misaka T, Arai S, Abe K. Gliadain, a gibberellin-inducible cysteine proteinase occurring in germinating seeds of wheat, Triticum aestivum L., specifically digests gliadin and is regulated by intrinsic cystatins. FEBS Journal . 2007;274:1908–1917. doi: 10.1111/j.1742-4658.2007.05749.x. [DOI] [PubMed] [Google Scholar]
- Koehler SM, Ho TH. Hormonal regulation, processing, and secretion of cysteine proteinases in barley aleurone layers. The Plant Cell. 1990;2:769–783. doi: 10.1105/tpc.2.8.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proceedings of the National Academy of Sciences, USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz MS, Ciborowski P. Zymographic techniques for detection and characterization of microbial proteases. Methods in Enzymology . 1994;235:563–594. doi: 10.1016/0076-6879(94)35171-6. [DOI] [PubMed] [Google Scholar]
- Leatherbarrow RJ. GraFit Version 7. Horley, UK: Erithacus Software Limited; 2009. [Google Scholar]
- Martinez DE, Bartoli CG, Grbic V, Guiamet JJ. Vacuolar cysteine proteases of wheat (Triticum aestivum L.) are common to leaf senescence induced by different factors. Journal of Experimental Botany. 2007;58:1099–1107. doi: 10.1093/jxb/erl270. [DOI] [PubMed] [Google Scholar]
- Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiology. 2009;151:1531–1545. doi: 10.1104/pp.109.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Cambra I, Gonzalez-Melendi P, Santamaria ME, Diaz I. C1A cysteine-proteases and their inhibitors in plants. Physiologia Plantarum. 2012;145:85–94. doi: 10.1111/j.1399-3054.2012.01569.x. [DOI] [PubMed] [Google Scholar]
- Martinez M, Diaz I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evolutionary Biology. 2008;8:198. doi: 10.1186/1471-2148-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Rubio-Somoza I, Carbonero P, Diaz I. A cathepsin B-like cysteine protease gene from Hordeum vulgare (gene CatB) induced by GA in aleurone cells is under circadian control in leaves. Journal of Experimental Botany . 2003;54:951–959. doi: 10.1093/jxb/erg099. [DOI] [PubMed] [Google Scholar]
- McLellan H, Gilroy EM, Yun BW, Birch PR, Loake GJ. Functional redundancy in the Arabidopsis cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytologist. 2009;183:408–418. doi: 10.1111/j.1469-8137.2009.02865.x. [DOI] [PubMed] [Google Scholar]
- Mikkonen A, Porali I, Cercos M, Ho TH. A major cysteine proteinase, EPB, in germinating barley seeds: structure of two intronless genes and regulation of expression. Plant Molecular Biology. 1996;31:239–254. doi: 10.1007/BF00021787. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Diaz I, Carrillo L, Fuentes R, Carbonero P. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. The Plant Journal. 2007;51:352–365. doi: 10.1111/j.1365-313X.2007.03146.x. [DOI] [PubMed] [Google Scholar]
- Parrott DL, Martin JM, Fischer AM. Analysis of barley (Hordeum vulgare) leaf senescence and protease gene expression: a family C1A cysteine protease is specifically induced under conditions characterized by high carbohydrate, but low to moderate nitrogen levels. New Phytologist . 2010;187:313–331. doi: 10.1111/j.1469-8137.2010.03278.x. [DOI] [PubMed] [Google Scholar]
- Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. The Plant Cell . 2000;12,:1031–1040. doi: 10.1105/tpc.12.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinlaor P, Kaewpitoon N, Laha T, et al. Cathepsin F cysteine protease of the human liver fluke, Opisthorchis viverrini . PLoS Neglected Tropical Diseases . 2009;3:e398. doi: 10.1371/journal.pntd.0000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research . 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends in Biochemical Sciences. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Schoberer J, Vavra U, Stadlmann J, Hawes C, Mach L, Steinkellner H, Strasser R. Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic. 2009;10:101–115. doi: 10.1111/j.1600-0854.2008.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Xu LL. Characters of cysteine endopeptidases in wheat endosperm during seed germination and subsequent seedling growth. Journal of Integrative Plant Biology. 2009;51:52–57. doi: 10.1111/j.1744-7909.2008.00778.x. [DOI] [PubMed] [Google Scholar]
- Shindo T, Misas-Villamil JC, Horger AC, Song J, van der Hoorn RA. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS One. 2012;7,:e29317. doi: 10.1371/journal.pone.0029317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T, Van der Hoorn RA. Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Molecular Plant Pathology. 2008;9:119–125. doi: 10.1111/j.1364-3703.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, et al. Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiology. 2008;146:1738–1758. doi: 10.1104/pp.107.111781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution . 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annual Review of Plant Biology . 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- Wang B, Shi GP, Yao PM, Li Z, Chapman HA, Bromme D. Human cathepsin F. Molecular cloning, functional expression, tissue localization, and enzymatic characterization. Journal of Biological Chemistry. 1998;273:32000–32008. doi: 10.1074/jbc.273.48.32000. [DOI] [PubMed] [Google Scholar]
- Weeda SM, Mohan Kumar GN, Richard Knowles N. Developmentally linked changes in proteases and protease inhibitors suggest a role for potato multicystatin in regulating protein content of potato tubers. Planta. 2009;230:73–84. doi: 10.1007/s00425-009-0928-0. [DOI] [PubMed] [Google Scholar]
- Wex T, Levy B, Wex H, Bromme D. Human cathepsins F and W: a new subgroup of cathepsins. Biochemical and Biophysical Research Communications. 1999;259,:401–407. doi: 10.1006/bbrc.1999.0700. [DOI] [PubMed] [Google Scholar]
- Wiederanders B, Kaulmann G, Schilling K. Functions of propeptide parts in cysteine proteases. Current Protein and Peptide Science. 2003;4:309–326. doi: 10.2174/1389203033487081. [DOI] [PubMed] [Google Scholar]
- Zhang N, Jones BL. Characterization of germinated barley endoproteolytic enzymes by two-dimensional gel electrophoresis. Journal of Cereal Science . 1995;21,:145–153. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.