Fig 2.
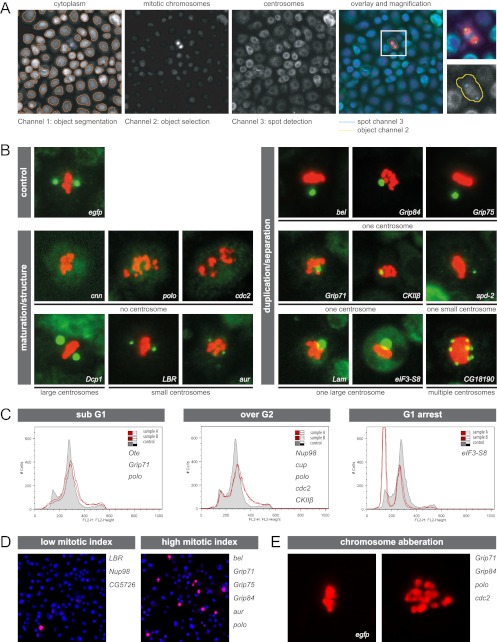
Functional characterization of phosphoproteins and regulatory kinases by RNAi, automated immunofluorescence microscopy, and FACS. (A) The image processing and segmentation approach, which was applied to assign identified phosphoproteins and kinases to regulatory pathways, is illustrated. The effects on number and size of mitotic centrosomes (labeled with anti-cnn, green) in RNAi-treated SL2 cells were analyzed using an algorithm that reports number, intensity, and morphology of intracellular objects. Cell cycle effects were monitored via calculating the ratio of total number of cells (labeled with cytoplasmic stain, blue) segmented in channel 1 to the number of mitotic cells (labeled with anti-phospho-histone H3, red) selected in channel 2. (B) Examples of mitotic cells (centrosomes in green and chromosomes in red) reflecting the 7 aberrant centrosome phenotypes observed after RNAi-mediated protein depletion: one centrosome, one large centrosome, one small centrosome, multiple centrosomes (duplication/separation); no centrosome (structure maintenance); and small and large centrosomes (maturation). Knockdown of EGFP served as a negative control. The corresponding RNAi target proteins are indicated within each image. (C) FACS analysis of SL2 cells incubated with dsRNA reveals genes involved in the regulation of cell cycle progression. Three types of aberrant cell cycle distribution profiles were identified: increased sub-G1 DNA content, increased number of polyploid cells, and accumulation of cells in G0/G1 phase. Each profile shown contains a control histogram (gray; cells treated with dsRNA targeting EGFP) and an aberrant histogram representative for its phenotypic class (red line; names of all target genes exhibiting similar profiles upon depletion are given in the corresponding panel). (D) Two fluorescence microscopy images (superimposition of DAPI [blue] and mitotic chromosomes [red]) are shown representative of cells displaying low or high mitotic indices after dsRNA treatment. Proteins whose depletion induced an aberrant proportion of mitotic cells are given on the right of the corresponding image. (E) Cells were manually scored for chromosome segregation defects after depletion of target proteins. A control cell treated with EGFP dsRNA with normal chromosome (red) alignment in metaphase and an example of abnormally distributed mitotic chromosomes are shown. The proteins inducing this phenotype are given on the right side of the panel.