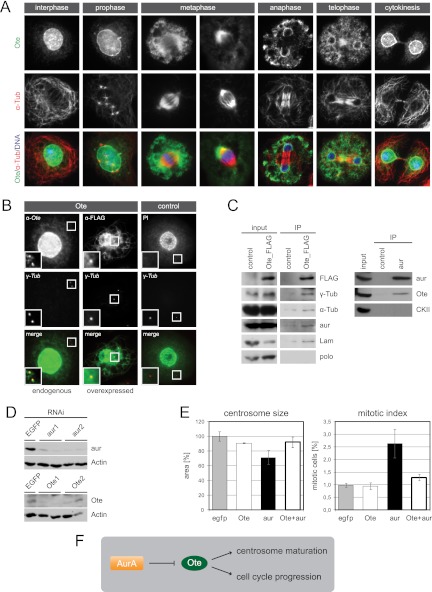
Fig 5.
Nuclear membrane protein Ote localizes to interphase centrosomes and functions downstream of aur in centrosome maturation. (A) Localization of endogenous Ote is shown throughout the cell cycle. SL2 cells were labeled with anti-Ote (green) and anti-α-Tub (red). Superimposition with DAPI in blue. (B) Endogenous Ote and overexpressed FLAG-tagged Ote localize to centrosomes in interphase. A fluorescence microscopy image of a cell labeled with rabbit anti-Ote (green) and anti-γ-Tub (red) is shown in the left panel. Cells stably expressing Ote fused to a FLAG tag were labeled with rabbit anti-FLAG (green) and anti-γ-Tub (red) (middle panel). The right panel shows a cell expressing TAP-tagged nuclear protein scra as a negative control, in which the tag was labeled with rabbit immunoglobulin (IgG, green) and the centrosome was labeled with anti-γ-Tub (red). Superimpositions of the two channels are shown in the bottom row, and magnifications of the area around the interphase centrosomes are given in each image. Fluorescent images in the right panel serve as a control for the specificity of the anti-Ote antibody and the specificity of fusion protein localization. Brightness and contrast of endogenous Ote staining were enhanced 3-fold in the magnified section to clarify localization of a minor portion of the protein to the centrosome while the majority of the protein localizes to the nuclear membrane. (C) Extracts from SL2 cells expressing FLAG-tagged Ote were analyzed by immunoprecipitation with anti-FLAG antibody. Interacting proteins were determined by Western blotting with antibodies to α-Tub, γ-Tub, aur, Lam, and polo. The known interaction of Lam and Ote was verified, and additionally, α-Tub, γ-Tub, and aur were found to copurify with FLAG-Ote while polo is not present in the purified complex. Immunoprecipitation of endogenous aur from Drosophila embryo homogenate verifies the interaction with Ote, which was detected in the aur precipitate but not in the control IP. Nontransfected cells served as control for the FLAG IP. (D) Western blots illustrating the efficiency of RNAi experiments. SL2 cells treated with two independent dsRNAs for each target gene show strongly decreased levels of aur and Ote, respectively, compared to control cells treated with dsRNA targeting EGFP. Actin is shown as a loading control. (E) Ote functionally interacts with aur. Phenotypes of single and double knockdowns regarding centrosome size and mitotic index are shown in the graphs. EGFP RNAi served as control. (F) The schematic illustrates the possible regulatory mechanism. Negative regulation of Ote by the kinase aur is indicated by ⊣.