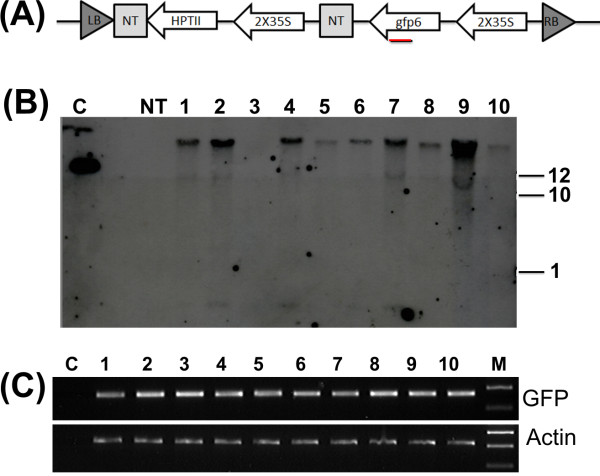
Figure 2.
Detection of transgenic hairy roots by Southern hybridization and Reverse Transcription PCR. (A) Schematic of GFP gene from pMDC44 showing the 35 S promoter, GFP gene, NOS terminator, HPT selection (Gateway Technologies Invitrogen, Carlsbad, CA, USA). Red bold line show the position used to amplify the GFP gene. Presence of GFP results in a 345 bp fragment. (B) Southern blot of transgenic roots. Lane 1 C is positive control (0.05 ng of pMDC44 plasmid), Lane 2 no sample loaded, Lane 3 NT non transformed wild type roots, lanes 4-13 consist of roots from 10 composite plants selected randomly (C) RT-PCR on transgenic and non-transgenic root cultures. The panel shows results obtained from using GFP specific primers and the same substrates amplified with Actin primers for loading control (lower panel). Lane 1 is 1 Kb ladder (Hyperladder I – Bioline) Lanes 2-11 cDNA Zea mays roots of composite plants, lane 12 cDNA from wild type formed maize roots.