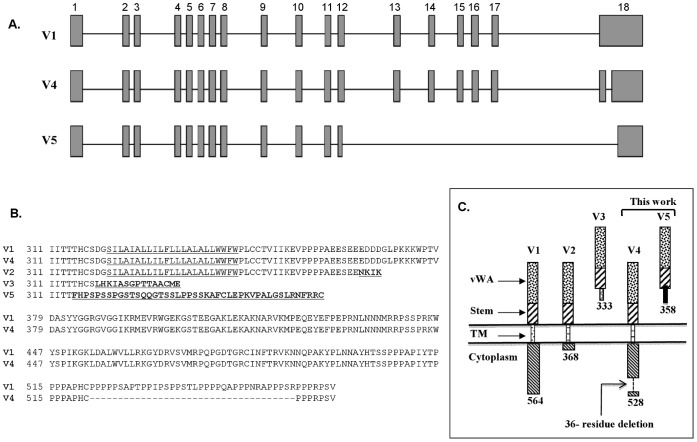
Figure 2. Comparison of ANTXR1/TEM8 variantsV1, V4 and V5 differential splicing.
A) Differential splicing of new variants V4 and V5 relative to V1. The gene comprises 18 exons with respect to V1 mRNA. V4 results from split splicing within exon 18, excluding a 108-bp segment, but it has the same stop codon as V1. V5 differential splicing occurs by partial skipping of exon 12 and 18 and complete skipping of exons 13–17. This results in a frame shift and consequently V5 acquires a new downstream stop codon. B) Sequence comparison of TEM8 receptor variants. The transmembrane helix (TM)) is underlined. The sequence differences are in underlined bold. V4 differs from V1 only in that it has a 36-residue in-frame deletion. V5, like V3, has no TM, and is therefore likely secreted. C) A schematic representation of membrane-bound and secreted variants of ANTXR1/TEM8. Same patterns signify identity of sequences and different ones divergence. The putative secreted variants V3 and V5 do not have any alternative membrane-spanning helices in their unique carboxyl-terminal segments.