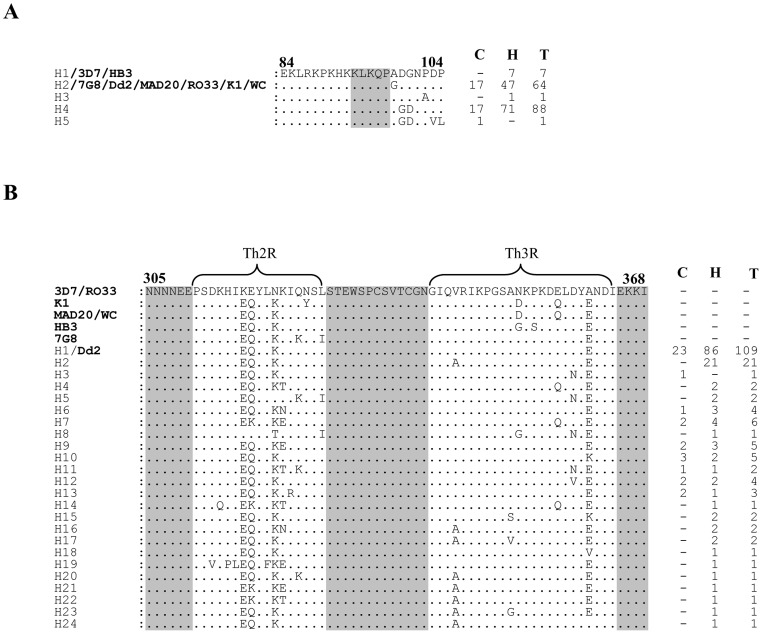
Figure 3. Sequence diversity in the N- and C-terminal non-repeat region of PfCSP.
(A) Sequence alignment showing polymorphisms in the non-repeat N-terminal T cell epitope region (amino acid residue 84 to 104) of CSP in 161 samples. The shaded region (amino acid residue 93 to 97) is a conserved motif involved in sporozoite invasion of mosquito salivary gland as well as in binding to hepatocytes prior to invasion. (B) Sequence alignment showing polymorphisms in the non-repeat C-terminal T cell epitope regions (Th2R spanning from amino acid residues 311 to 327 and Th3R from amino acid residues 341 to 364) of CSP in 179 samples. The highly conserved sequences flanking the Th2R and Th3R domains are shaded grey. The eight laboratory-adapted strains are also included in this alignment. Numbers on the right indicate numbers of samples belonging to that particular haplotype. Dots represent amino acid positions identical to the 3D7 haplotype, whereas those different are indicated. C, community cohort; H, hospital cohort; T, total; WC, Wellcome.