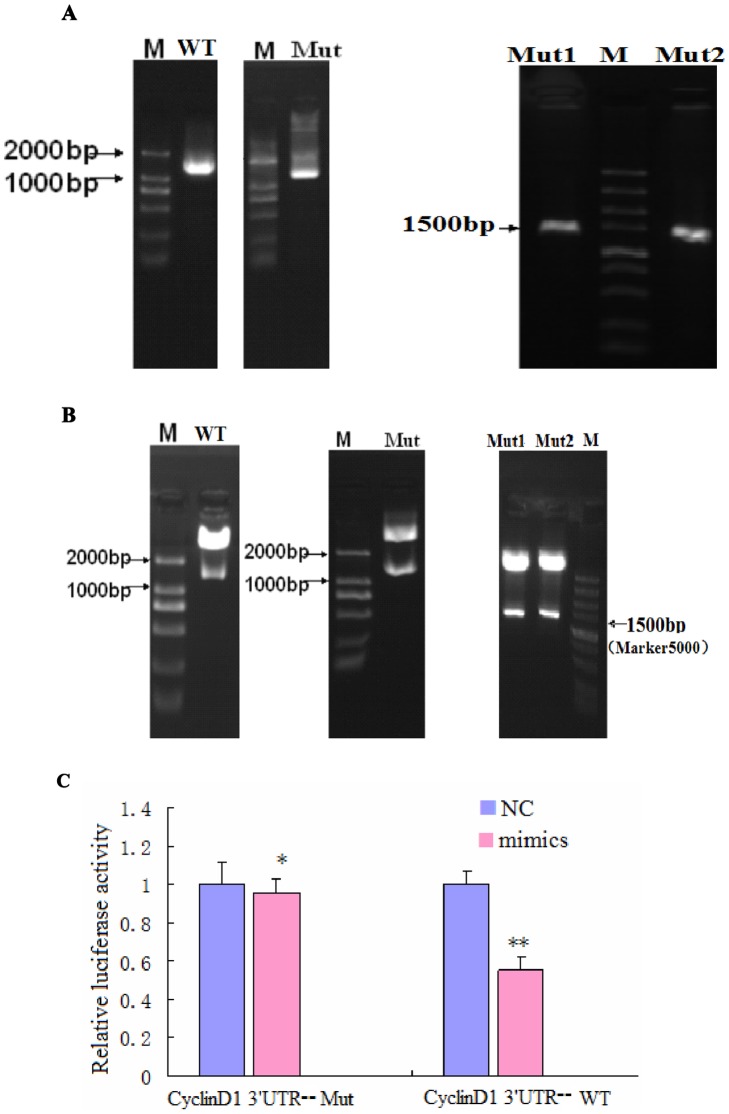
Figure 6. CyclinD1 is a direct target of and is regulated by miR-338-3p, and the effect of miR-338-3p on CyclinD1 is mainly dependent on the CyclinD1-3′-UTR region (nt 2397–2403).
(A–B) Successfully constructed plasmids of CyclinD1-3′-UTR. (A) Amplification of a DNA fragment containing CyclinD1-3′-UTR region. PCR amplification was performed using specific primers for the types of WT-, Mut-, Mut1-, and Mut2-Cyclin D1-3′-UTR, respectively, and genomic DNA isolated from the genome of LO2/HBx cells was used as PCR template. An approximately 1573 bp band PCR product was detected by agarose gel electrophoresis. (B) Identification of recombinant plasmids. The fragment digested with XhoI and NotI from the recombinant plasmids: pCyclinD1-3′-UTR-WT, pCyclinD1-3′-UTR-Mut, pCyclinD1-3′-UTR-Mut1, and pCyclinD1-3′-UTR-Mut2 were used as a template for confirmation PCR. The PCR product was about 1573 bp, which was finally confirmed by DNA sequencing. miR-338-3p targets CyclinD1. (C) Dual luciferase assay of LO2/HBx cells cotransfected with the Renilla luciferase constructs containing the CyclinD1 WT or Mut 3′-UTR and miR-338-3p mimics or negative RNA (*P = 0.404,**P<0.001). (D) The major site of CyclinD1 targeted by miR-338-3p was at position 2397–2403 nt. Effects of miR-338-3p and the negative control on the reporter constructs containing CyclinD1-3′-UTR Mut1 and Mut2 were determined 48 hours after transfection. Renilla luciferase values normalized to Firefly luciferase are presented. (**P<0.001,***P = 0.04). (E–G) CyclinD1 protein expression after transfection of miR-338-3p mimics, inhibitor, or negative control (*P<0.001,**P<0.01). The fold change before and after transfection is more pronounced in LO2/HBx-d382 cells than that in LO2/HBx and control cells. Data are shown as mean ±SD from at least 3 independent experiments. (H) qRT-PCR with the 2−ΔΔCT method to evaluate the CyclinD1 mRNA expression normalized to GAPDH in LO2/HBx-d382, LO2/HBx, and control cells transfected with miR-338-3p mimics or inhibitor or their respective controls(★P = 0.164,★★ P = 0.438, #P = 0.220,##P = 0.101,* P = 0.254,** P = 0.417).