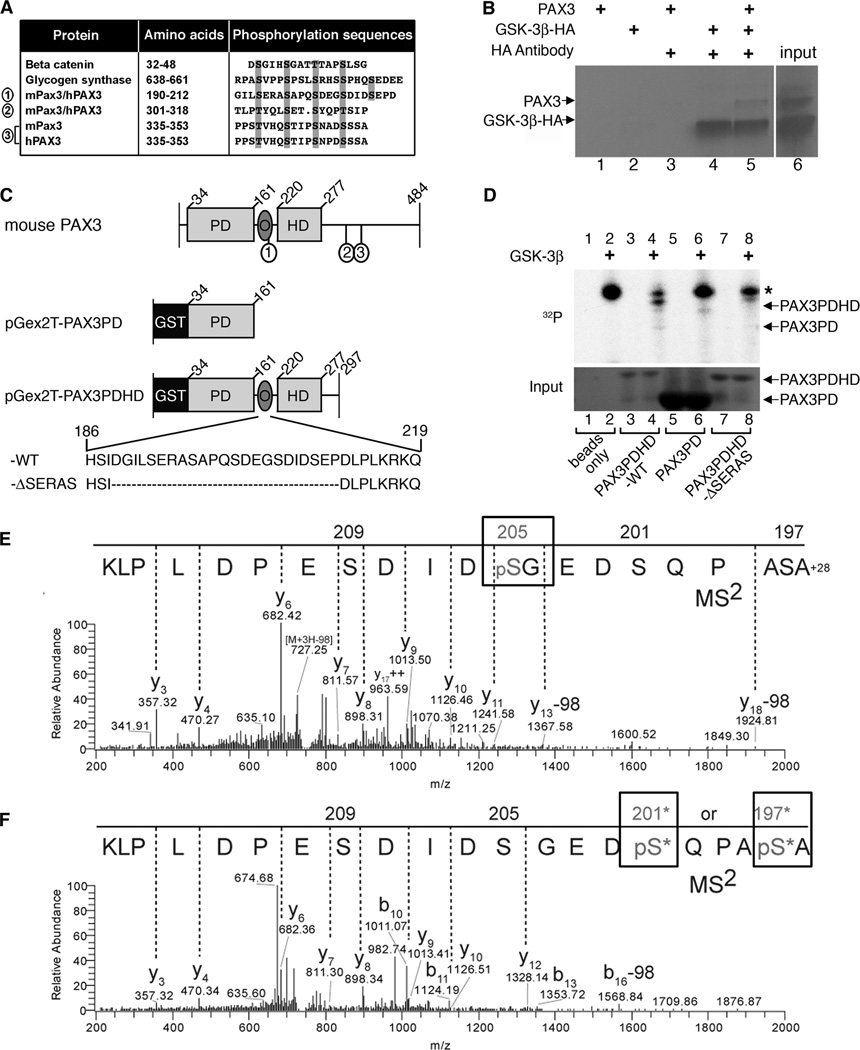
Figure 6.
GSK-3β interacts with and phosphorylates PAX3. A, PAX3 possesses three conserved putative GSK-3β triplicate recognition motifs (sites 1, 2, 3) which are aligned with known GSK-3β targets β-catenin and glycogen synthase. B, immunoprecipitation of GSK-3β and PAX3. Sepharose-A/G beads were mixed with radiolabeled PAX3 (lane-1), HA-GSK-3β (lane-2), PAX3 with HA antibody (lane-3), HA-GSK-3β with HA antibody (lane-4) and both PAX3 and HA-GSK-3β with HA antibody (lane-5). Input of radiolabeled PAX3 and HA-GSK-3β is shown in lane-6. C, schematic of recombinant GST-tagged murine PAX3 proteins. Full-length wild type mouse PAX3 depicts placement of the three putative GSK-3β recognition motifs (1, 2, 3) corresponding to sites in A. The pGex2T-PAX3PD construct fuses glutathione S-transferase (GST) to the N-terminal end of the paired domain (PD) and contains amino acids 34–161. pGex2T-PAX3PDHD-WT possesses amino acids 34–297 including the PD, octapeptide (O) and the homeodomain (HD). The amino acid sequence of O and the first GSK-3β recognition motif (S/T-X3-S/T) is represented (wild-type (WT) sequence shown, amino acids 186–219). The construct pGex2T-PAX3PDHD-ΔSERAS has the entire series of GSK-3β recognition motifs removed (deleted sequence, (−ΔSERAS) sequence shown, with the removal of amino acids 189–211). D, GSK-3β and PAX3 kinase assay. The kinase assay was performed on empty glutathione sepharose-4B beads without (lane-1) or with GSK-3β (lane-2), pGex2T-PAX3PDHD-WT without (lane-3) or with GSK-3β (lane-4), pGex2T-PAX3PD without (lane-5) or with GSK-3β (lane-6) and pGex2T-PAX3PDHD-ΔSERAS minus (lane-7) or plus GSK-3β (lane-8). The top panel displays the kinase assay with an asterisk indicating an auto-phosphorylation band. The bottom panel is a coomassie-stained gel visualizing the input bound to the beads. E–F, tandem mass spectrometry determines Ser205 and Ser197/Ser201 of PAX3 are phosphorylated by GSK-3β in-vitro. Precursor ion masses were measured in the Orbitrap analyzer and MS2 spectra were acquired in the LTQ mass spectrometer. E, MS2 spectra of pSer205 (n-formyl) ASAPQSDEGpSDIDSEPDLPLK (MS mass deviation, 12 ppm). F, MS2 spectra of peptide AS*APQS*DEGSDIDSEPDLPLK phosphorylated at either Ser197 or Ser201 (MS mass deviation, 23 ppm). The presence of ion Y12 at mass 1328.14m/z in the Y-ion series fragmentation of the MS2 spectra exclude Ser209 and Ser205 at the phosphorylation site, however, there is insufficient MS2 ion evidence (b1–9) to pinpoint the phosphorylation site specifically to Ser197 or Ser201, thus both sites with an * are potential sites of phosphorylation. Note: peptide is displayed C- to N-terminus due to the predominant Y-ion fragmentation.