Figure 1. Features of epimutations likely vary in complexity, necessitating sophisticated experimental designs and possibly explaining why some designs fail.
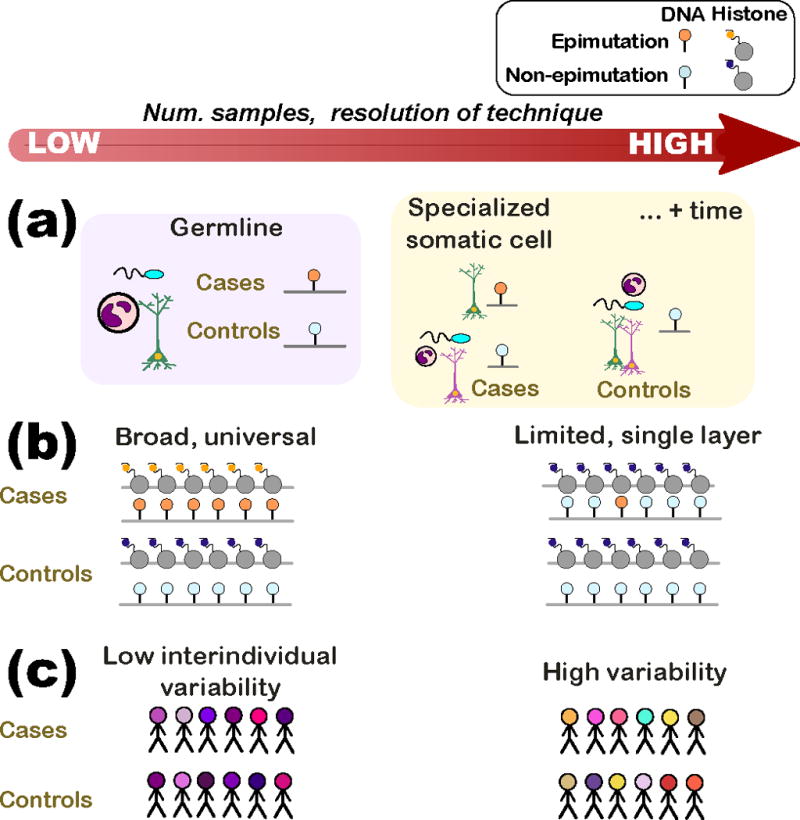
(a) An epimutation may be inherited (germline) or may be acquired during the individual’s lifetime. In the former case (purple box), an epimutation would be consistently present in all cell types of the affected individuals. In the latter (orange box), the epimutation may only be found in particular cell types of the affected individual or, still more complex, in a specialized cell type following a particular developmental stage (e.g. after puberty).
(b) Epimutations may vary in the stretch of epigenome affected and in the number of epigenetic layers where the mutation is evident. (Left) An epimutation that spans a large stretch of the epigenome (similar to a translocation) and multiple epigenetic layers would be detected by a low-resolution technique and several assays. (Right) At the other extreme, an epimutation that affects a single locus (similar to a point mutation) in a single layer would require single base-pair resolution techniques and would not be found in assays investigating other epigenetic modifications.
(c) The interindividual variability in the epigenome surrounding the epimutation also affects sample size and technique resolution. (Left) Regions of the epigenome that have low interindividual variability (e.g. conserved regulatory domains) require fewer samples than in cases where the epimutation is in a region of high variability (i.e. regulatory region affected by lifestyle choices). There is currently little understanding of the population variability of the epigenome.
