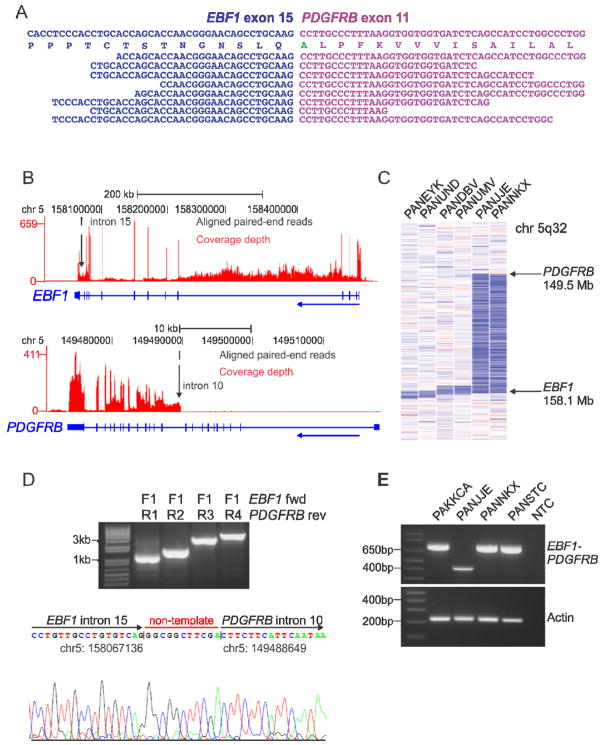
Figure 3.
mRNA-seq data, recurrence screening and genomic mapping of the EBF1-PDGFRB fusion. (A) Split reads mapping across the EBF1-PDGFRB fusion point for case PAKKCA. Amino acid substitution from wild-type PDGFRB (Ser>Ala), is highlighted in green. (B) Coverage depth for all mRNA-seq reads at the EBF1 and PDGFRB locus in case PAKKCA, showing expression across the EBF1 locus and increased expression of PDGFRB at intron 10 (arrowed). The vertical height of the red bar indicates the number of reads covering the site. (C) SNP 6.0 microarray log2 ratio DNA copy number heatmap showing deletion (blue) between EBF1 and PDGFRB for two EBF1-PDGFRB cases (PANJJE and PANNKX), and four non-rearranged cases with focal EBF1 deletions (left). (D) Genomic mapping of the EBF1-PDGFRB rearrangement breakpoint by PCR (top) and sequencing (bottom), showing juxtaposition of EBF1 intron 15 (chr5:158067136) to PDGFRB intron 10 (chr5:149488649), with the addition of non-template nucleotides between the breakpoints. (E) RT-PCR confirmation of EBF1-PDGFRB fusion in four high-risk B-ALL cases with exon 14 (bottom band) or exon 15 (top band) of EBF1 fused to exon 11 of PDGFRB. See also Figure S3.