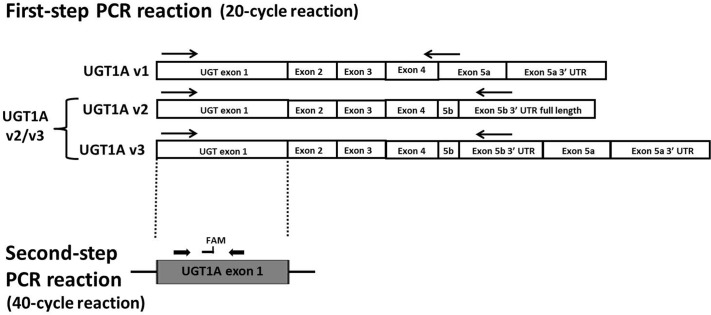
Fig. 1.
Schematic of UGT1A expression analysis. The first-step PCR amplification involved a 20-cycle reaction to distinguish between the different exon 5 splice variants. Because UGT1A v1 variants encode the active i1 proteins, whereas UGT1A v2 and v3 variants encode the same inactive i2 proteins, UGT1A v2 and v3 were grouped together and designated UGT1A v2/v3 mRNA as described previously (Girard et al., 2007). The arrows represent the approximate location of the primers, with the same antisense primer used for amplification of both UGT1A v2 and v3 mRNA. UGT1A exon 1 is representative of the nine different UGT1A exon 1 species that can be alternatively spliced to the common UGT1A exons 2 to 5. The second-step real-time PCR assay was then performed by using UGT1A exon 1-specific real-time PCR primers and probes. FAM, approximate location of fluorescent 6-carboxyfluorescein probe.