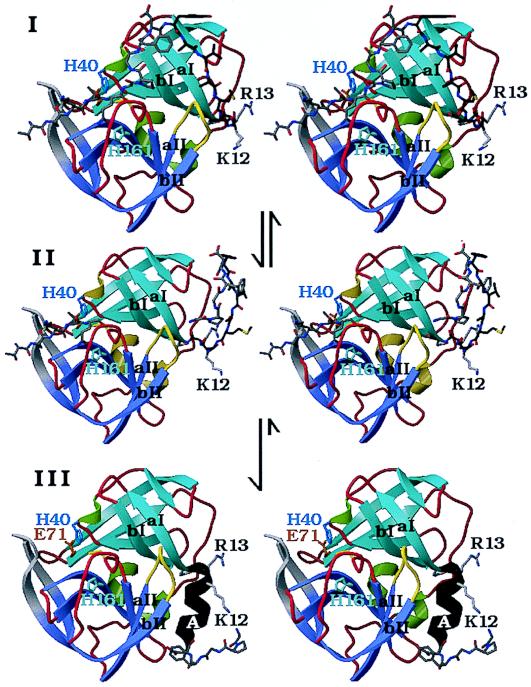
Figure 5.
A structural model for the N-terminal, autocatalytic excision of the enteroviral 3C proteases. The model is based on the crystal structure of the poliovirus 3C protease (HPV 3C) (63). The secondary structure of HPV 3C is shown in a ribbon representation. The N-terminal β-barrel domain is blue and the C-terminal β-barrel domain is mauve. (I) Model of the precursor of the 3C protease with the 3B|3C cleavage site sequence bound in the active site in the conformation of a cognate substrate of the 3C protease. The model was derived from the P5 to P2′ residues of OMTKY3 bound in the active site of Streptomyces griseus protease B (SGPB) (65) after the optimal superposition of HPV 3C and SGPB (63). Included in this figure are the residues starting at the P5 (Thr) position of the 3B protein. The P5 to P1 residues and residues 1–5 of HPV 3C (P1′ to P5′) are colored gray; residues 6–13 are dark gray. The side chains of three active site residues, the nucleophile Cys-147 (yellow), the general acid-base catalyst His-40 (blue), and the S1 specificity determinant His 161 (light blue), are included. Residues 1–11 of HPV 3C reach into the active site of the protease and are in a mostly extended conformation. After the intramolecular cleavage the new N terminus Gly-1 dissociates from the active site while the P5 to P1 residues are still bound (II). Subsequently residues 6–13 of HPV 3C fold into a stable α-helix (colored black), which prevents the new N terminus from binding again to the active site and renders the conformational change irreversible. Arg-13 of the conserved sequence motif K/RR/KNL/I, which forms the last turn of the N-terminal helix in HPV 3C, anchors the N terminus to the structure. (III) The crystal structure of HPV 3C is shown with the N-terminal α-helix in black. The rearrangement of the N terminus in this model is accompanied by small conformational changes of β-strands aI and bI of the N-terminal domain and the loop (yellow) that connects β-strands aII and bII of the C-terminal domain.