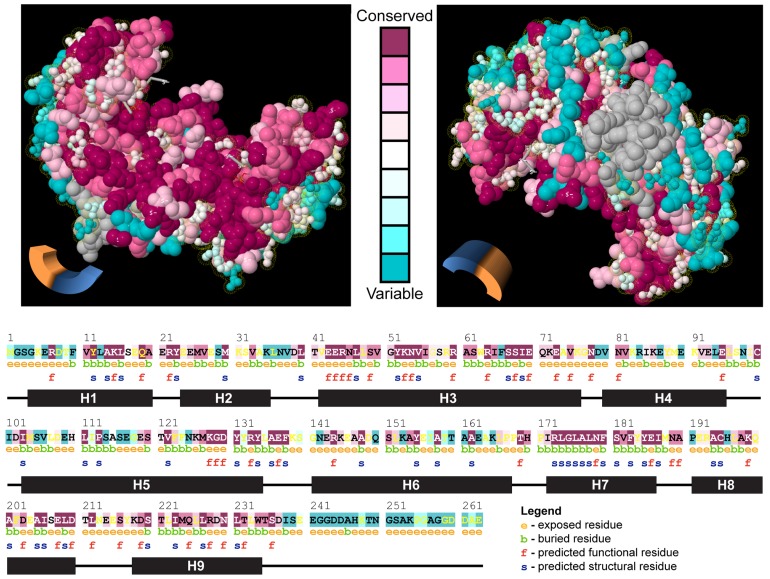
FIGURE 3.
Amino acid conservation among the 13 Arabidopsis 14-3-3 isoforms. The Amino acid conservation among the 13 Arabidopsis 14-3-3 isoforms is mapped onto the 14-3-3 protein structure (based on that of human 14-3-3τ/θ) using ConSurf (Landau et al., 2005; Ashkenazy et al., 2010; top panel). Color-coding indicates the amount of change each position has undergone over evolutionary time. Views are angled with perspectives shown in lower left corners, with monomers in orange and blue. Residue conservation is highest in the client binding channel, which is exposed in the left image. The consensus 14-3-3 amino acid sequence is color-coded according to the degree of conservation determined with ConSurf (bottom panel). Residues predicted to have roles in 14-3-3 structure (conserved and buried) and function (conserved and exposed) are annotated below the sequence. Exposed and buried residues were determined using a neural network algorithm. Residues for which there was insufficient data (present in less than 10% of isoforms) are indicated in yellow. Positions of the nine alpha-helices are indicated below the sequence.