Abstract
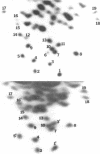
Nucleotide sequences of large T1 ribonuclease fragments of 18S ribosomal RNA of Novikoff rat ascites hepatoma cells and chicken lymphoblastoid cells were determined and compared. Among the 19 large T1 ribonuclease fragments examined of rat 18S ribosomal RNA, 12 fragments were found to be the same in chicken 18S ribosomal RNA. Three fragments of rat 18S ribosomal RNA were not found among large T1 ribonuclease fragments of chicken 18S ribosomal RNA. Four fragments of rat 18S ribosomal RNA were found to be changed in chicken 18S ribosomal RNA. All the changes were point mutations except the change in the largest T1 ribonuclease fragment 1 which is 21 nucleotides long. 2'-0-methylation at the center of the fragment was lost in chicken 18S ribosomal RNA; all the other nucleotides were the same.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendich A. J., McCarthy B. J. Ribosomal RNA homologies among distantly related organisms. Proc Natl Acad Sci U S A. 1970 Feb;65(2):349–356. doi: 10.1073/pnas.65.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Fuke M., Busch H. A T1 ribonuclease fragment present in 18 S ribosomal RNA of Novikoff rat ascites hepatoma cells and absent from 18 S ribosomal RNA of HeLa cells. J Mol Biol. 1975 Dec 5;99(2):277–281. doi: 10.1016/s0022-2836(75)80145-8. [DOI] [PubMed] [Google Scholar]
- Fuke M., Busch H. Partial methylation of 18 S ribosomal RNA detected by T1 ribonuclease digestion and homochromatography fingerprinting. FEBS Lett. 1977 May 15;77(2):287–290. doi: 10.1016/0014-5793(77)80253-6. [DOI] [PubMed] [Google Scholar]
- Fuke M., Busch H., Rao P. N. Evolutionary trends in 18S ribosomal RNA nucleotide sequences of rat, mouse, hamster and man. Nucleic Acids Res. 1976 Nov;3(11):2939–2957. doi: 10.1093/nar/3.11.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke M., Busch H. Sequence analysis of T1 ribonuclease fragments of 18S ribosomal RNA by 5'-terminal labeling, partial digestion, and homochromatography fingerprinting. Nucleic Acids Res. 1977 Feb;4(2):339–352. doi: 10.1093/nar/4.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S. A. Fine structure of ribosomal RNA. I. Conservation of homologous regions within ribosomal RNA of eukaryotes. J Mol Biol. 1976 Sep 25;106(3):791–816. doi: 10.1016/0022-2836(76)90265-5. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequences within the ribosomal ribonucleic acids of HeLa cells, Xenopus laevis and chick embryo fibroblasts. J Mol Biol. 1976 Feb 25;101(2):235–254. doi: 10.1016/0022-2836(76)90375-2. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Forbes J., de Jonge P., Klootwijk J. Presence of a hypermodified nucleotide in HeLa cell 18 S and Saccharomyces carlsbergensis 17 S ribosomal RNAs. FEBS Lett. 1975 Nov 1;59(1):60–63. doi: 10.1016/0014-5793(75)80341-3. [DOI] [PubMed] [Google Scholar]
- Padgett T. G., Stubbledield E., Varmus H. E. Chicken macrochromosomes contain an endogenous provirus and microchromosomes contain sequences related to the transforming gene of ASV. Cell. 1977 Apr;10(4):649–657. doi: 10.1016/0092-8674(77)90098-8. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Jeppesen P. G. Extent of variation in three related bacteriophage RNA molecules. J Mol Biol. 1972 Jul 28;68(3):417–428. doi: 10.1016/0022-2836(72)90096-4. [DOI] [PubMed] [Google Scholar]