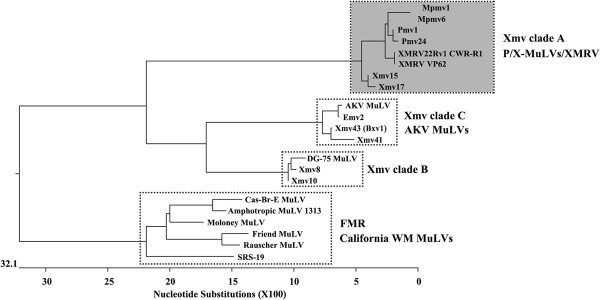
Figure 7.
Phylogenetic analysis of glyco-gag nucleotide sequences in MuLVs. The nucleotide sequences in the glyco-gag region for endogenous MuLVs (nt 1–264, for M-MuLV J02255 nt 357 – 620) were aligned by DNASTAR Lasergene 8 MegAlign (Clustal W method) and a phylogenetic tree is shown. The same regions from representative exogenous MuLVs of the Friend-Moloney-Rauscher (FMR) group are also aligned. Box with the dark shading indicates endogenous MuLVs that do not have sequences capable of encoding glyco-gag. Boxes without shading contain viruses or endogenous MuLVs that can encode glyco-gag.