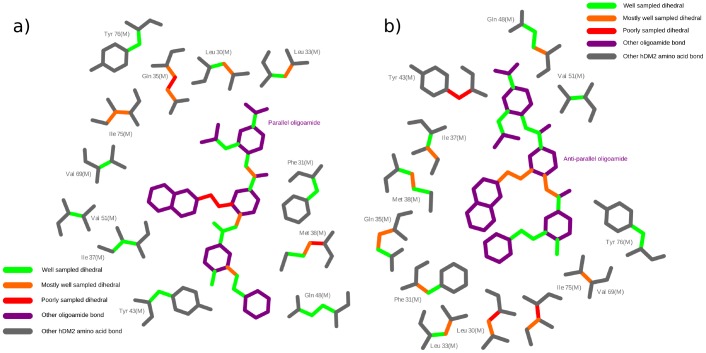
Figure 8. Dihedral sampling of binding site hDM2 residues and arylamide bonds.
2D representations of a) parallel and b) anti-parallel conformations of the Phe-Nap-iPr arylamide in the hDM2 binding site are produced using Ligplot [51]. Amino acid backbone residues and bonds with no dihedrals are shown in grey, while arylamide torsions without flexible dihedrals are shown in purple. Rotatable bonds are colored according to our sampling criteria with: green (well sampled); orange (well sampled in all but one simulation); red (poorly sampled across simulations). It is clear that many dihedral angles are well sampled, but some are not, indicating that multiple starting configurations must be used for calculating thermodynamic properties of the system.