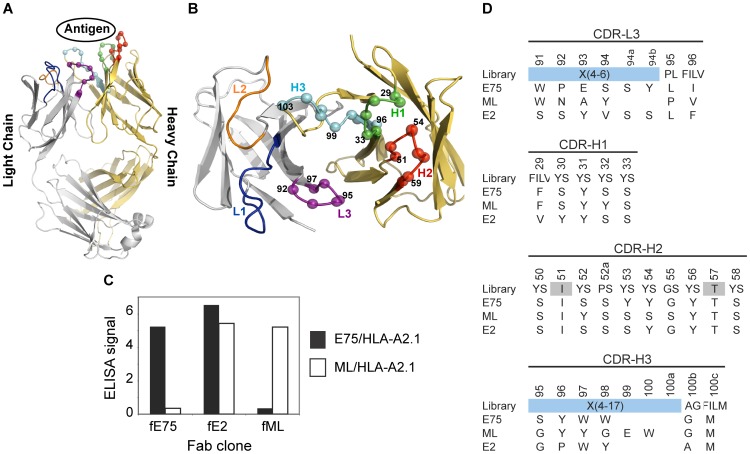
Figure 1. Phage-Display isolation of Fabs Specific for pMHC Molecules.
Fabs specific for either E75/HLA-A2 or ML/HLA-A2 molecules were selected by phage-display technology. Each Fab construct was built from the highly stable 4D5 Fab scaffold containing both a heavy and light chain each with a single variable and constant domain (modeled from PDB ID: 1FVD) (A). The diversity of the Complementary Determining Regions (CDR) for the heavy chain; H1, H2, and H3, and the light chain; L3, was restricted in favor of tyrosine, serine, and other small amino acids. The Cα atoms of the synthetically modified H1, H2, H3, and L3 regions are shown as spheres (B). After three rounds of selection, an ELISA was used to test the specificity of the amplified clones (C). Three clones with three different specificities were identified. The Fab clones fE75 and fML bound E75/HLA-A2 and ML/HLA-A2 respectively with no detectable binding to the opposite pMHC molecule. The Fab clone fE2 showed binding to both E75/HLA-A2 and ML-HLA-A2. Following each Fab clone's specificity determination, the amino acid sequence of each clone was determined (D).