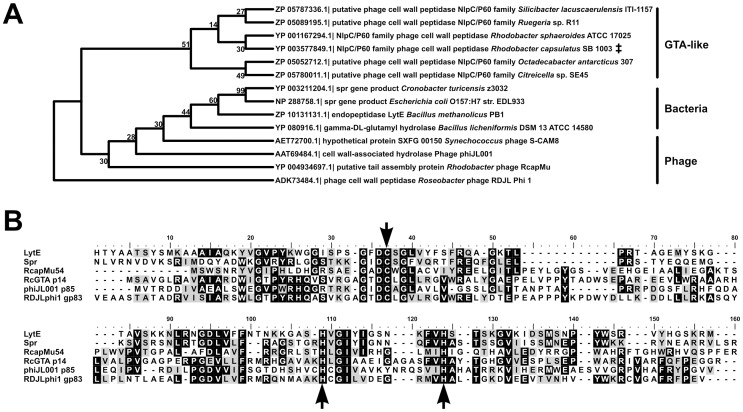
Figure 7. Bioinformatic analysis of RcGTA g14.
A. Maximum parsimony phylogenetic analysis of the five closest Blastp hits to a p14 query, the two top Spr protein hits, the two top LytE hits, and the four highest scoring bacteriophage hits. Database accession numbers are given on the left, and annotations are on the right. The RcGTA p14 protein is indicated (‡). Bootstrap values result from 1,000 iterations. B. MUSCLE alignment [56] of RcGTA p14 protein with Spr (E. coli O157:H7 EDL933), LytE (Bacillus methanolicus PB1), ФJL001 p85, RDJLФ1 gp83 and RcapMu54. Conserved catalytic residues are indicated by arrows. The threshold for shading is 50%; black represents identity and grey represents similarity according to the BLOSUM matrix. Alignment tails with no sequence identity were clipped.