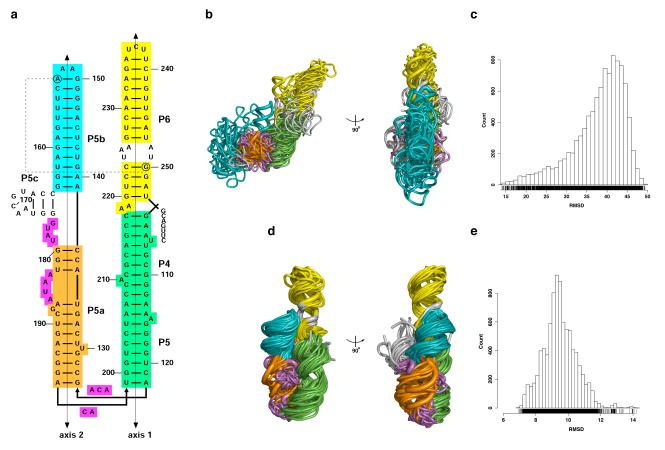
Figure 3. P4-P6 conformational sampling.
(Top) Low set. a) Secondary structure coloured by stems: P4-P5 (green); P5a (orange); P5b (cyan); and, P6 (yellow). The other nucleotides are in magenta. The nucleotides that were not sampled are uncoloured. b) Two views of twenty centroids (thin tubes) that are optimally aligned on the crystal structure (PDB file 1GID, thick tube). The colours are the same as in a. c) RMSD (Å) range (all-atoms) of the low set models computed against the P4-P6 crystal structure (PDB code 1GID). (Bottom) High set. The A-minor long-range base pair (A153-G250) is added in the constraints (circled nucleotides and dashed line). d) Two views of twenty centroids (thin tubes) that are optimally aligned on the crystal structure (PDB file 1GID, thick tube). The colours are the same as in a. e) RMSD (Å) range (all-atoms) of the high set models computed against the P4-P6 crystal structure (PDB code 1GID).