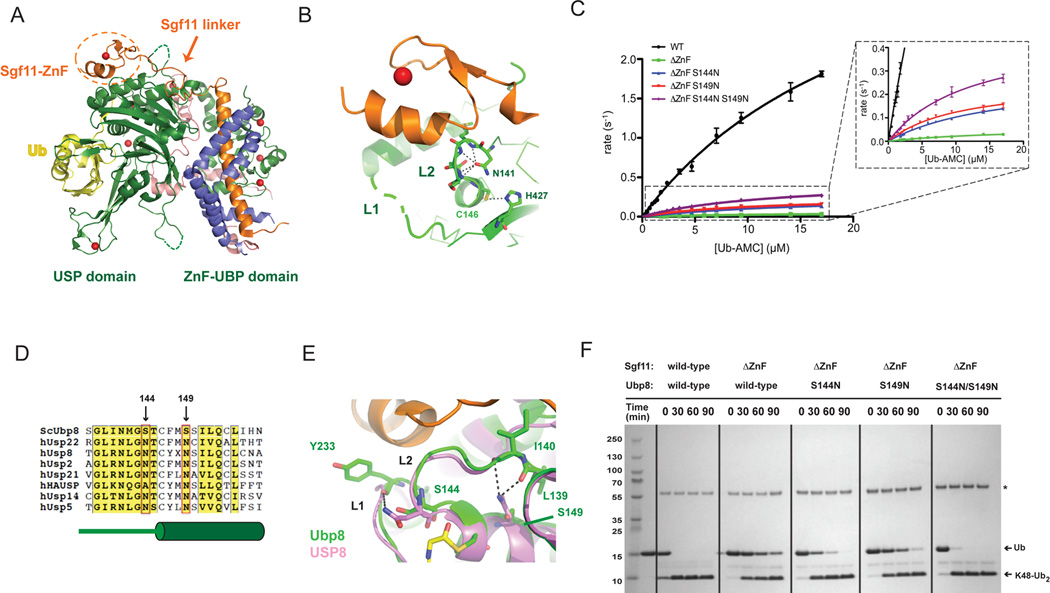
Figure 1. Gain-of-function mutations in Ubp8 that compensate for deletion of the Sgf11 zinc finger.
(A) Structure of the wild-type DUBm. The four subunits are Ubp8 (green), Sgf11 (orange), Sus1 (blue), Sgf73 (salmon); zinc atoms are depicted as red spheres. The USP and ZnF-UBP domains of Ubp8 are indicated below. The Sgf11-ZnF (in dotted circle) binds to the USP domain of Ubp8 and is connected to its helical region by an extended linker.
(B) The Sgf11-ZnF (orange) binds near the Ubp8 (green) active site, contacting loops L1 and L2. The catalytic residues, C146 and H427, and the oxyanion hole residue, N131 are depicted as sticks. Loop L1 (dashed gray line) is disordered in the absence of ubiquitin.
(C) Rate of Ub-AMC cleavage as a function of substrate concentration by wild-type DUBm and DUBm lacking the Sgf11-ZnF containing either wild-type or Ubp8 with substitutions S144N, S149N, or S144N/S149.
(D) Sequence alignment of USP DUBs indicated residues 144 and 149 of Ubp8. Diagram in green below sequence indicates secondary structure in Ubp8.
(E) Superposition of Ubp8 (green) with hUSP8 (pink) showing hydrogen bonding between N149 in hUSP8 with loop L2 backbone residues. Residue S149 in Ubp8 does not contact loop L2.
(F) Cleavage of K48-diubiquitin by wild-type and mutant DUBm. Gel stained with Coomassie.
See also Figure S2.