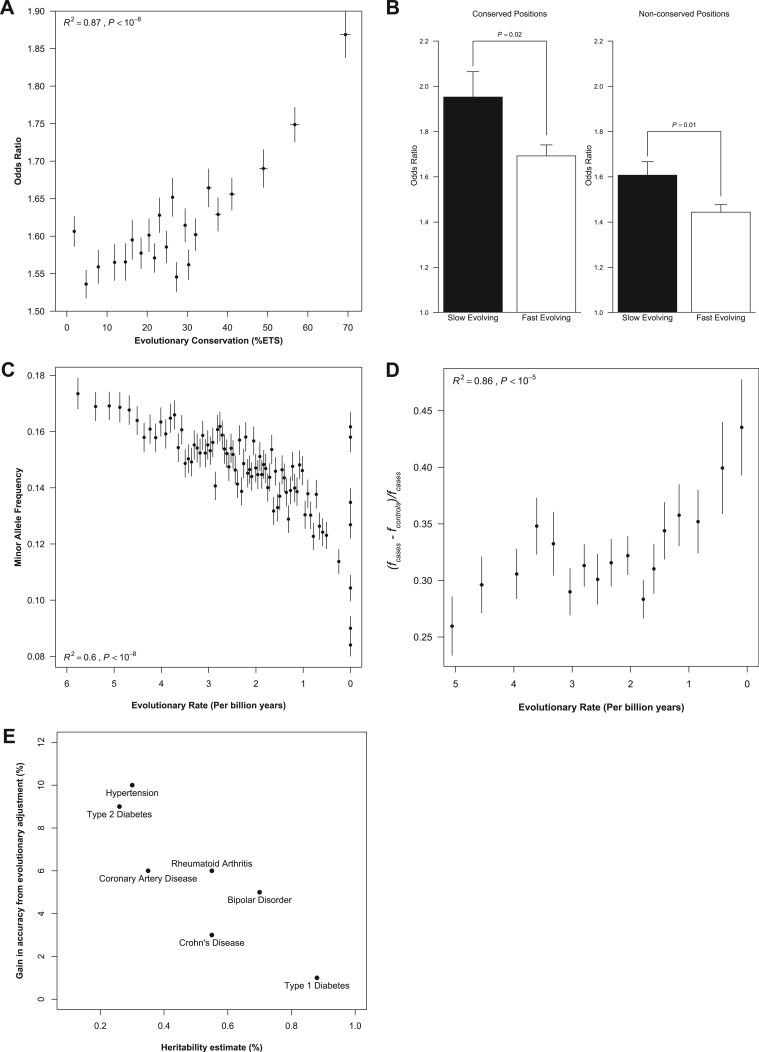
FIG. 2.
The effect size of disease variants relates to the evolutionary anatomy of genomic positions. (A) Relationship between the odds ratio reported for disease-associated variants and the evolutionary conservation of the genomic position harboring the variant. All reported odds ratios were normalized toward disease risk estimation by taking the exponent of the absolute loge of odds ratios < 1. Each point represents the mean of nonoverlapping bins of n = 200 associated loci ordered by increasing %ETS. The trend was best described by a second order polynomial (R2 = 0.87, P < 10−8; Pearson r = 0.86, P < 10−4) (error bars: standard error of mean [SEM]). (B) Slowly evolving sites (bottom 25% of evolutionary rates) at highly conserved positions (top 25% of evolutionary conservation) exhibit higher average odds ratios than faster-evolving sites (top 25% of evolutionary rates) at both conserved (top 25% ETS) and nonconserved (bottom 25% ETS) positions (error bars: SEM). (C) Relationship of the multispecies evolutionary rate with the MAFs in human populations. Each point is estimated as the average of evolutionary rate and MAFs for 100,000 SNPs randomly sampled from HapMap 3 CEU population data (second order polynomial R2 = 0.6, P < 10−8; Pearson r = −0.76; P < 10−4) (error bars: SEM). (D) The influence of evolutionary rate on the risk allele frequency disparities between cases and controls. Δf is the difference in risk allele frequency between cases (fcases) and controls (fcontrols) divided by fcontrols to control for the MAF of the risk allele in healthy populations; Δf = (fcases − fcontrols)/ fcontrols. Each point represents the mean of nonoverlapping bins of n = 200 associated loci ordered by increasing evolutionary rate (third order polynomial R2 = 0.86, P < 10−5; Pearson r = 0.74, P < 10−3) (error bars: SEM). (E) For each of the diseases measured in the WTCCC, the gain in predictive accuracy (i.e., difference between the evolutionary adjusted P value (E-rank) AUC and the “raw” P value (P-rank) AUC) is plotted against the total heritability estimate for the disease (Sofaer 1993; Poulsen et al. 1999; Katzmarzyk et al. 2000; Smoller and Finn 2003; Agarwal et al. 2005; Harney et al. 2008; Ounissi-Benkalha and Polychronakos 2008).