FIG. 6.
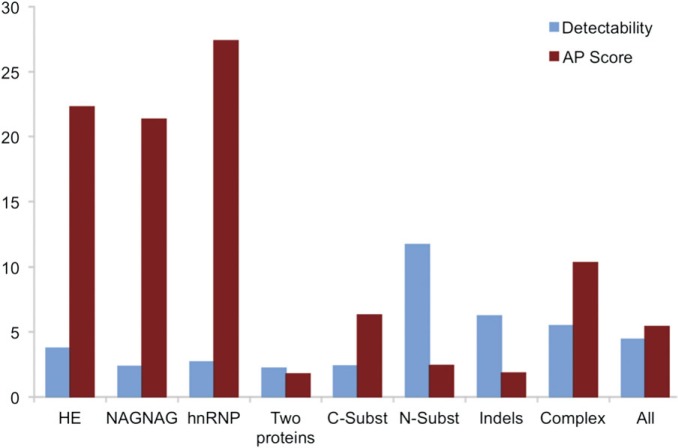
Alternative isoforms generated from three subsets are supported by more peptide evidence. The percentage of PSM that distinguish alternative isoforms (the AP Score) contrasted with the theoretical likelihood of detecting splice isoforms (detectability) calculated for each of the eight subsets from the AI Detected set. Detectability is calculated from the number of PSM detected for the each gene and the theoretical probability of detection, both normalized by the equivalent values from the Background set. The higher the detectability, the more likely we are to detect PSM that match to alternative isoforms. The AP Score is surprisingly high for three of the groups (the hnRNP genes, the genes with isoforms generated from NAGNAG splicing, and the genes with alternative isoforms generated from homologous exons).