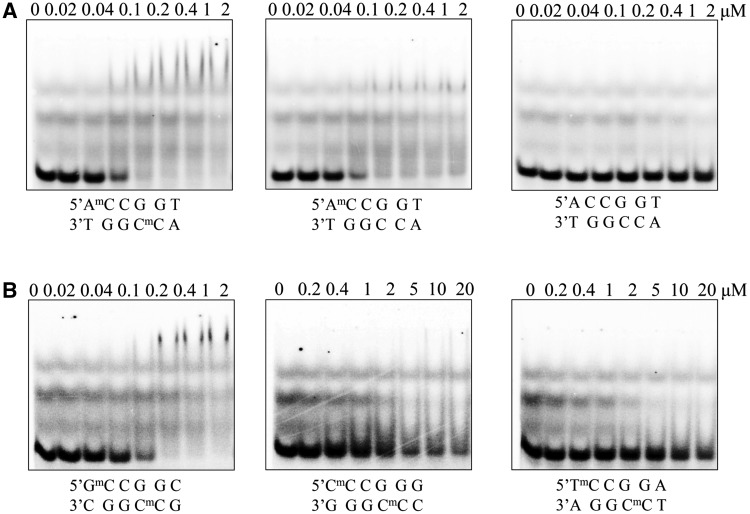
Figure 2.
Gel mobility shift analysis for McrB-N binding to DNA. (A) McrB-N binding to the oligoduplexes used for crystallization. McrB-N at concentrations indicated above the relevant lanes was mixed with the di-methylated (m/m), hemi-methylated (m/−) or non-methylated (−/−) DNA (Figure 1B) at the final concentration of 100 nM. The samples were electrophoresed through 8% PAA gels under native conditions. (B) Dependence of the McrB-N binding on the sequence context 5′-upstream of 5mC McrB-N at concentrations indicated above the relevant lanes was mixed with the di-methylated (m/m) DNA containing either G, C or T nucleotide upstream of the 5mC. The samples were analysed as described in (A).