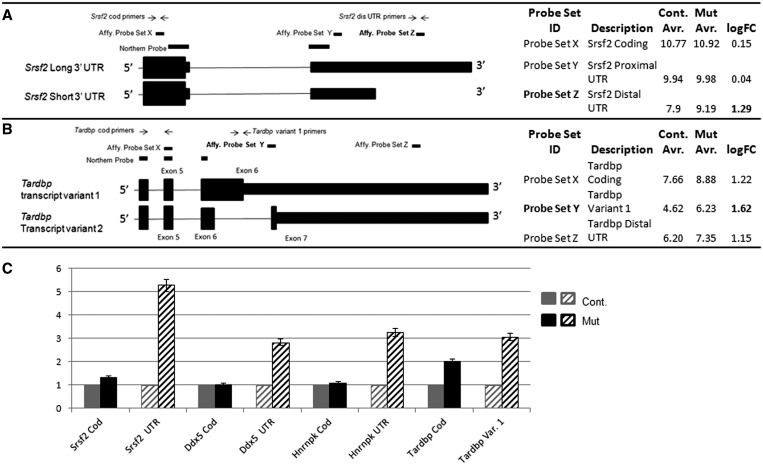
Figure 2.
Detailed examination of Affymetrix probe sets yields unexpected information about mRNA processing as well as expected changes in total transcription. (A and B) Cartoon schematics of two genes, Srsf2 and Tardbp, showing the location of the Affymetrix probes, designed qPCR primers and northern blot probes. The qPCR primer location mirrors the positions of the Affymetrix probes, and northern blot probes are chosen to hybridize to all transcripts. (A) Three Affymetrix probes to the Srsf2 gene are depicted: one in the coding region, one in the proximal 3′-UTR and one in the distal 3′-UTR. The probe in the distal 3′-UTR would uniquely hybridize to the long transcript of Srsf2. In control spermatids, hybridization of the Srsf2 distal UTR probe is low compared to the coding and proximal UTR probes demonstrating lack of the Srsf2 long transcript. The long transcript is present in mutant spermatids. (B) Three Affymetrix probes to the Tardbp gene are depicted: one in the coding region, one unique to transcript variant 1 and one in the distal 3′-UTR. Tardbp transcription is up-regulated in mutant spermatids, but the possibility exists that there may be a specific up-regulation of transcript variant 1. (C) Q-PCR of four selected genes. Overall levels of transcription of Srsf2, Ddx5 and Hnrnpk are not altered in mutant round spermatids. However, the presence of normally truncated 3′-UTR sequence is increased. In contrast, overall levels of Tardbp transcription are increased in mutant round spermatids, in particular, for transcript variant 1.