Abstract
Although most DNA polymerases discriminate against ribonucleotide triphosphaets (rNTPs) during DNA synthesis, recent studies have shown that large numbers of ribonucleotides are incorporated into the eukaryotic nuclear genome. Here, we investigate how a DNA polymerase can stably incorporate an rNTP. The X-ray crystal structure of a variant of human DNA polymerase λ reveals that the rNTP occupies the nucleotide binding pocket without distortion of the active site, despite an unfavorable interaction between the 2′-O and Tyr505 backbone carbonyl. This indicates an energetically unstable binding state for the rNTP, stabilized by additional protein–nucleotide interactions. Supporting this idea is the 200-fold lower catalytic efficiency for rNTP relative to deoxyribonucleotide triphosphate (dNTP) incorporation, reflecting a higher apparent Km value for the rNTP. Furthermore, distortion observed in the structure of the post-catalytic product complex suggests that once the bond between the α- and β-phosphates of the rNTP is broken, the unfavorable binding state of the ribonucleotide cannot be maintained. Finally, structural and biochemical evaluation of dNTP insertion onto an ribonucleotide monophosphate (rNMP)-terminated primer indicates that a primer-terminal rNMP does not impede extension. The results are relevant to how ribonucleotides are incorporated into DNA in vivo, during replication and during repair, perhaps especially in non-proliferating cells when rNTP:dNTP ratios are high.
INTRODUCTION
The stability of the genome depends on the fidelity of synthesis by DNA polymerases. To ensure accurate DNA replication and repair at each incorporation step, DNA polymerases have to select a nucleotide substrate containing a correct base and a correct sugar. Incorporation of a substrate with an incorrect sugar, i.e. a ribonucleotide (rNTP), may be the source of genomic instability. Even a single rNMP residue in DNA may stall a DNA polymerase and arrest replication. Furthermore, the 2′-OH on the ribose ring may promote hydrolytic cleavage of the DNA backbone. Most DNA polymerases select against ribonucleotide incorporation via steric exclusion of the 2′-OH of the ribose (1). A crystal structure of the ternary complex of a family Y Dpo4 Y12A mutant with an incoming adenosine diphosphate, suggests that in the wild-type Dpo4, the side chain of Tyr12 would cause a steric clash with the 2′-OH of the incoming rNTP (2). Similarly, in high fidelity polymerases from families A and B, a bulky side chain of a residue at the nucleotide binding pocket has been identified as a ‘steric gate’ for preventing ribonucleotide incorporation (3,4). Nevertheless, even the high fidelity replicative polymerases occasionally incorporate ribonucleotides despite their relatively high sugar selectivity, defined as the ratio of catalytic efficiencies for deoxyribonucleotide triphosphate (dNTP) to ribonucleotide triphosphaets (rNTP) incorporation. Recent studies of the yeast replicative polymerases (Pols α, δ and ε) estimate that more than 10 000 rNTPs may be stably incorporated into the genome during one round of replication (5,6). These studies suggest that rNTPs may be the most common non-canonical nucleotides incorporated into the eukaryotic genome. The probability of incidental rNTP incorporation is likely increased by the fact that cellular rNTP levels exceed those of dNTPs by 10- to 200-fold. It has been suggested that the erroneously incorporated rNTPs can be excised by concerted actions of RNase H type 2 and Flap endonuclease-1 (FEN-1) (7,8).
rNTPs may not always be incorporated inadvertently; in some cases especially during repair reactions in non-proliferating cells where the relative concentrations of rNTPs are higher compared to that of dNTPs, they may serve as legitimate substrates. It has been suggested that, during DNA double-strand break repair (DSBR) by non-homologous end joining (NHEJ), family X polymerases participating in this repair pathway could incorporate rNTPs (9–11). In contrast to high fidelity polymerases from families A and B, family X polymerases (in mammalian cells: Pols λ, β, µ and Terminal deoxynucleotidyl transferase (TdT)) have relatively low sugar selectivity and thus more frequently incorporate rNTPs. However, even among members of this family, sugar selectivity differs by several orders of magnitude. Mammalian TdT and Pol µ exhibit the lowest sugar selectivity, with values ranging from 1 to 10 (9–11). The sugar selectivity of yeast Saccharomyces cerevisiae Pol IV is also relatively low (values on the order of 10–50) (12). Mammalian Pols β and λ are more proficient in sugar discrimination, with sugar selectivity values between 2 × 103 and 8 × 103 (13,14). Nonetheless, even the higher sugar selectivity of Pols β and λ remains significantly lower than that of replicative polymerases of family A and B (3,4).
Recent studies suggest that family X polymerases do not rely on a bulky side chain of a residue at the nucleotide binding pocket for steric exclusion of the 2′-OH on the ribose. Rather, family X polymerases appear to exclude rNTPs due to the steric clash of the 2′-OH with the peptide backbone of a residue at the nucleotide binding site. Crystal structures of the ternary complexes of wild-type and Y271A human Pol β, with an incoming rNTP (15), provide an initial understanding of the binding event of an rNTP to Pol β. The aim of the current structural study is to gain further understanding of how DNA polymerases discriminate against rNTPs and how ribonucleotides are stably incorporated in spite of sugar discrimination.
The variant of human Pol λ, Pol λ DL, used for this study was obtained by replacing nine residues from Loop 1 with a shorter four-residue sequence from human Pol β, as described earlier (16). This modification reduced the base substitution fidelity of the variant enzyme without a significant change of the catalytic efficiency for correct insertion. It also did not alter the geometry of the active site for correct incorporation. In fact, the only noticeable difference between the structures of the pre-catalytic ternary complexes of Pol λ DL and wild-type (wt) Pol λ with a dNTP, is in the Loop 1 region (16). We suggested that the modification of Loop 1 eliminated one (or more) of the kinetic checkpoints that prevent misincorporation, lowering the free energy required to achieve active site geometry compatible with catalysis (17). Therefore, we hypothesized that similar to reduced discrimination against base–base mismatches, rNTP discrimination by Pol λ DL may be also reduced, making Pol λ DL a good model for structural studies of rare events such as rNTP incorporation. Indeed, while our initial attempts to obtain a crystal structure of the wt Pol λ with rNTP were not successful, we were able to obtain crystal structures of Pol λ DL with rNTP. The four structures discussed here represent distinct steps in the catalytic cycle for rNTP incorporation (binding, insertion and extension, Figure 1). They reveal, how a ribonucleotide can be accommodated in the polymerase active site and how an rNMP-terminated primer can be extended resulting in a stable ribonucleotide incorporation into the genome during DNA replication and repair.
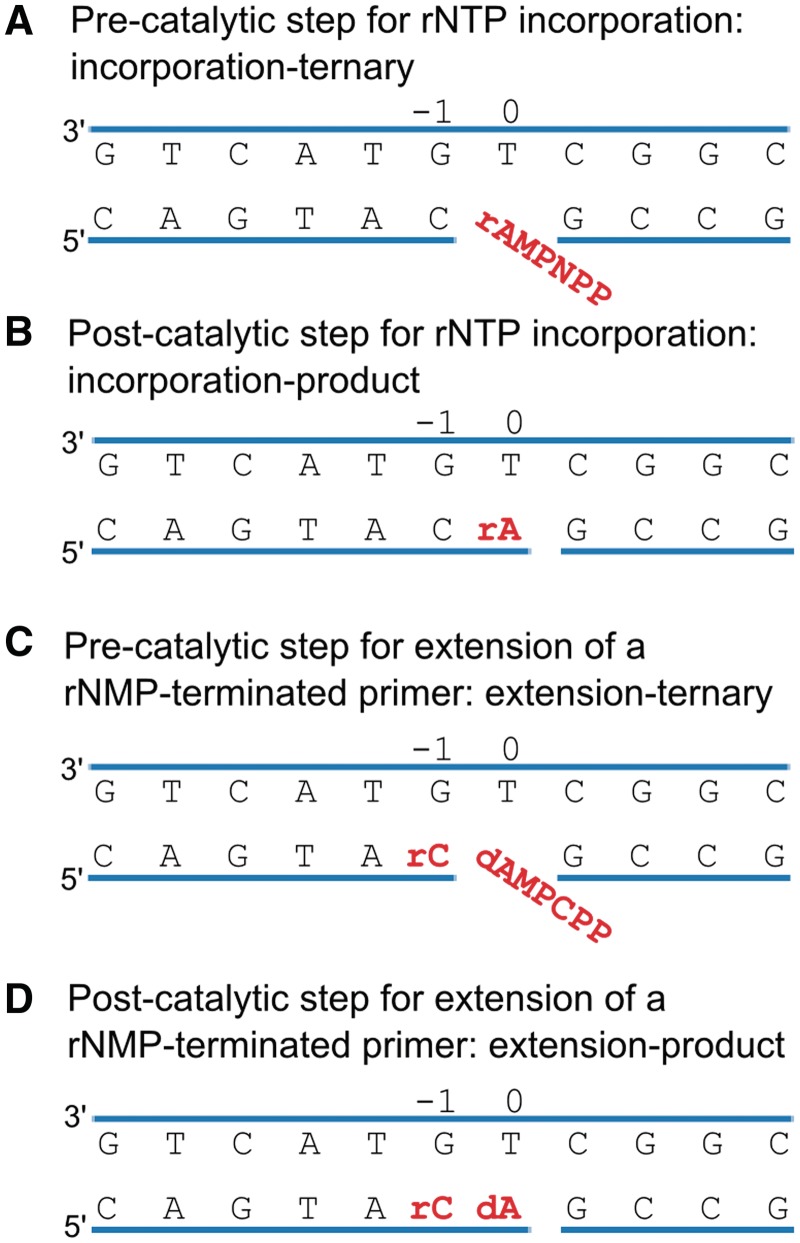
Figure 1.
Schematic depicting the four steps in the catalytic cycle of Pol λ DL. (A) Pre-catalytic complex for rNTP incorporation: incorporation-ternary. The bound rAMPNPP is highlighted in red. (B) Post-catalytic complex for rNTP incorporation: incorporation-product. The inserted rAMP is highlighted in red. (C) Pre-catalytic complex for extension of a rNMP-terminated primer: extension-ternary. Both the inserted rCMP and the bound dAMPCPP are highlighted in red. (D) Post-catalytic complex for extension of a rNMP-terminated primer: extension-product. The two inserted nucleotides are highlighted in red. Nucleotide base pairs at position −1 and 0 sites are indicated.
MATERIALS AND METHODS
Materials
The 39-kDa polymerase domain of human Pol λ loop variant (Pol λ DL) was expressed in Escherichia coli and purified as described in Supplementary Methods. Oligonucleotides were from Oligos Etc. (Wilsonville, OR, USA). The C543A mutation was introduced to prevent intermolecular disulfide bond formation during crystallization (18).
Steady-state kinetics
The steady-state measurements of single nucleotide incorporation were performed as described elsewhere (16). DNA substrates were prepared by hybridizing a 32P-5′-end-labeled 17-nucleotide primer (P17T, 5′-GTACGACTGAGCAGTAC or P17rC, 5′-GTACGACTGAGCAGTArC) and a 14-nucleotide downstream primer (DPT, 5′-GCCGGACGACGGAG) with a phosphate on the 5′-end to a 32-mer template (T32GT, 5′-CTCCGTCGTCCGGCTGTACTGCTCAGTCGTAC) to create a one-nucleotide gap substrate. Reaction mixtures (20 µl) contained 50 mM Tris, pH 7.5, 1 mM dithiothreitol (DTT), 4% (v/v) glycerol, 0.1 mg/ml bovine serum albumin, 2.5 mM MgCl2, 200 nM DNA and 39 kDa domain of wt Pol λ or Pol λ DL. Reactions were initiated by adding deoxyribose adenosine triphosphate (dATP) or adenosine triphosphate (rATP) at concentrations indicated below and incubated at 37°C. To measure incorporation of dATP onto a primer-terminal deoxyribose cytosine monophosphate (dCMP), reaction mixtures contained 1 or 1.5 nM of wt Pol λ or 1.5 nM of Pol λ DL as well as one of the following concentrations of the dATP: 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5 and 10 µM, and the reactions were incubated for 3 min. For measuring incorporation of rATP, each reaction mixture contained 10 nM Pol λ DL or wt, and one of the nine concentrations of rATP (10, 20, 30, 45, 60, 100, 150, 250 and 350 µM) or the reaction contained 3 nM Pol λ DL and one of the nine concentrations of rATP (1, 2, 5, 10, 20, 40, 60, 70 and 90 µM). The reactions were incubated at 37°C for 6 or 3 min, for wt and Pol λ DL, respectively. To measure extension from a primer-terminal cytosine monophosphate (rCMP), each reaction contained 2 nM wt Pol λ and one of nine concentrations of dATP (0.01, 0.02, 0.05, 0.2, 0.5, 1, 3, 6 and 12 µM) or reactions contained 5 nM Pol λ DL and dATP at one of the ten concentrations (0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 3, 6 and 12 µM). The reactions mixtures were incubated at 37°C for 3 min. After adding an equal volume of loading dye [99% (v/v) formamide, 5 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromophenol blue], products were resolved on a 16% denaturing polyacrylamide gel and quantified by phosphor screen autoradiography. The data were fit to the Michaelis–Menten equation using non-linear regression from KaleidaGraph software version 3.6.
Crystallography
Pre-catalytic complex for ribonucleotide incorporation (incorporation-ternary)
Oligonucleotides T11T (5′-CGGCTGTACTG), PG1 (5′-CAGTAC) and DT (5′-GCCG) were mixed in equimolar ratios in 100 mM Tris pH 7.5 and 40 mM MgCl2. The DNA was annealed in a thermal cycler by denaturation at 94°C, followed by a slow temperature gradient from 90 to 4°C. Hybridized DNA was mixed with 16 mg/ml Pol λ DL (3:1 DNA/protein ratio) at 4°C for 1.5 h before adding adenosine-5′-[(α,β)-imido]triphosphate (rAMPNPP) to 0.9 mM. The mixture was incubated at 4°C for 1 h. Protein/DNA mixture (1 µl) was combined with 1 µl of 100 mM Na HEPES pH 7.5 and 1.0 M Li2SO4. Crystals were grown at 20°C using hanging-drop vapor diffusion. Crystals were transferred in three steps to a solution containing 100 mM Na HEPES pH 7.5, 1.0 M Li2SO4, 100 mM MnCl2, 63 mM NaCl, 1 mM DTT, 0.9 mM rAMPNPP and 15% (v/v) ethylene glycol.
Post-catalytic complex for ribonucleotide incorporation (incorporation-product)
Crystals for pre-catalytic ternary complex grown as described above were used to obtain the structure of post-catalytic complex. These crystals were transferred in three steps to a solution containing 100 mM Na HEPES pH 7.5, 1.0 M Li2SO4, 100 mM MnCl2, 63 mM NaCl, 1 mM DTT, 15% (v/v) ethylene glycol and 10 mM rATP. The crystals were soaked overnight to ensure exchange of the rAMPNPP and incorporation-turnover of the rATP.
Pre-catalytic complex for extension of a rNMP-terminated primer (extension-ternary)
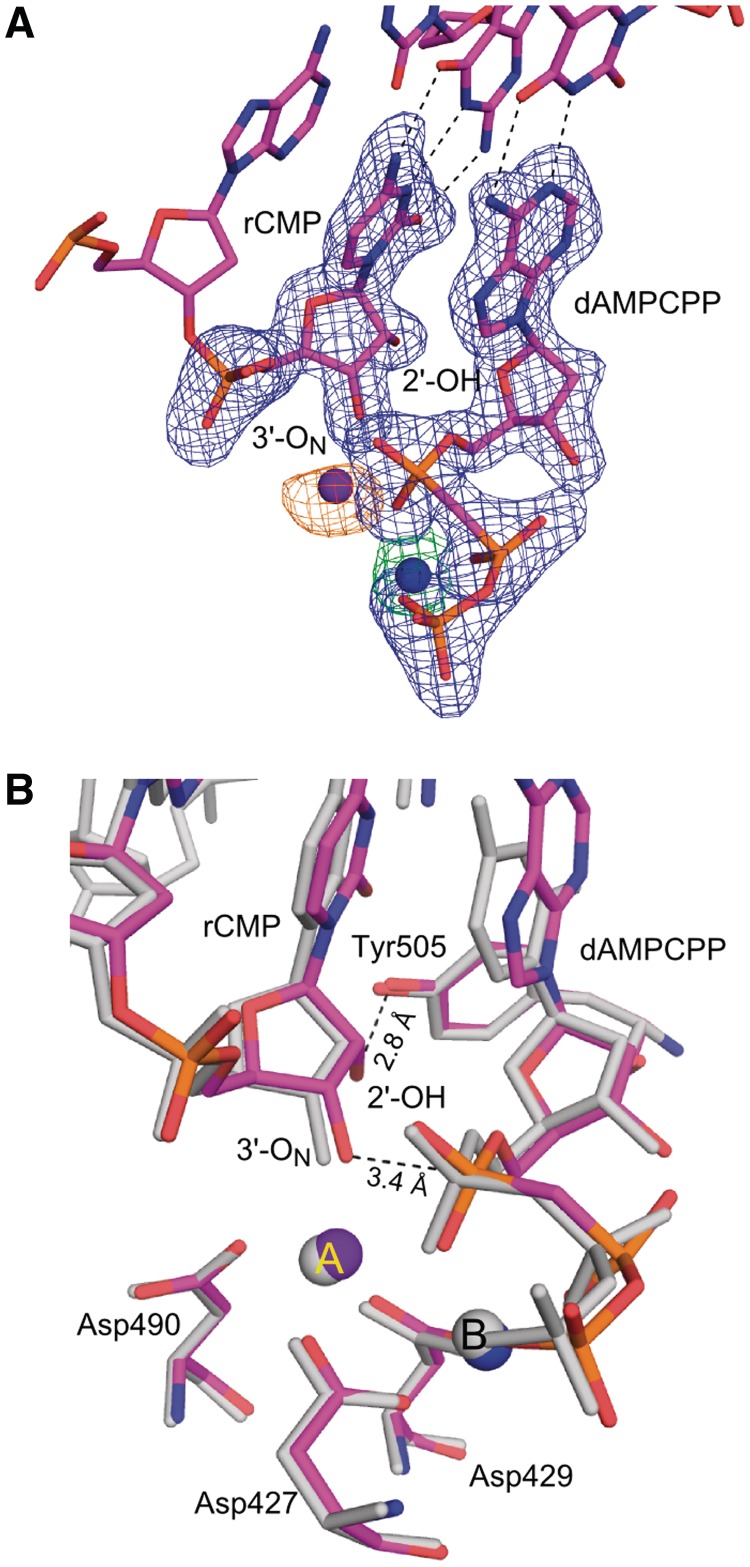
Crystals of Pol λ DL in complex with oligos T11T, PG2 (5′-CAGTA), DT, cytosine triphosphate (rCTP) and 2′-Deoxy-adenosine-5′-[(α,β)-methyleno]triphosphate (dAMPCPP) were grown using hanging-drop method. Protein/DNA mixture was prepared as described above. After incubation at 4°C for 1.5 h, 10 mM rCTP and 0.9 mM dAMPCPP were added. The mixture was incubated at 4°C for 1 h. These crystals grew at 20°C in 50 mM Na MES pH 6.5, 9% (v/v) PEG 400, 100 mM KCl and 10 mM MgCl2. Crystals were transferred in three steps to a solution containing 50 mM Na MES pH 6.5, 15% (v/v) PEG 400, 100 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 63 mM NaCl, 17.5% (v/v) glycerol and 1 mM dAMPCPP. The crystallization of this complex includes an additional molecule of DNA bound between two molecules of Pol λ DL at a crystallographic interface. We have modeled a 5-mer duplex DNA in the density consistent with the presence of excess primer/template duplex DNA prior to incorporation of the rCMP on the primer (Supplementary Figure S3). The presence of this extraneous DNA results in movement in two adjacent loop regions; residues 406–415 and 432–444 which are slightly displaced as compared to those in structures lacking the extra DNA. The presence of this DNA molecule causes no apparent changes in the active site.
Post-catalytic complex for extension of a rNMP-terminated primer (extension-product)
Crystals of Pol λ DL in complex with oligos T11T, PG2, DT, rCTP and dATP were grown using the sitting drop method. rCTP and dATP were added at 10 mM concentrations to the protein/DNA mixture prepared as described above. The mixture was incubated at 4°C for 1 h. These crystals grew at 20°C in 170 mM ammonium acetate, 85 mM sodium citrate tribasic dihydrate pH 5.6, 25.5% (w/v) PEG 4000, 15% (v/v) glycerol and were directly flash frozen.
All crystals were flash frozen in liquid nitrogen and data collected between −170 and −180°C. All data were indexed and scaled using the HKL2000 data processing software (19). The structures were solved using the structure of ternary complex of Pol λ DL [Protein Data Bank (PDB) ID code 3MGI, (16)] as a starting model. Reference Rfree reflections were maintained. The structures were obtained by iterative cycles of refinement and model building using Phenix (20) and Coot (21). Model quality was analyzed using Molprobity (22).
RESULTS
Steady-state kinetic assays for rNTP incorporation and extension
We first examined the proficiency of Pol λ DL to incorporate rNTPs. To this end, we measured the catalytic efficiencies of wt Pol λ and Pol λ DL for rATP incorporation using a steady-state kinetic assay. Because stable incorporation depends on the efficiency of extension of the incorporated nucleotide, we also measured the extension efficiency of an rNMP-terminated primer by both polymerases (Table 1 and Figure 1A and C). The catalytic efficiency of Pol λ DL was similar to that of wt Pol λ for correct incorporation of dATP, but it was 19-fold higher for incorporation of rATP. Pol λ DL showed a 16-fold lower discrimination factor for rATP incorporation relative to that of the wt enzyme. This suggested that Pol λ DL incorporated rNTPs more efficiently than the wt Pol λ. Both Pol λ DL and wt incorporated a correct dATP onto a rCMP-terminated primer with similar catalytic efficiencies as for incorporation onto a dCMP-terminated primer. Thus, discrimination factors for extension of a rNMP-terminated primer were similar for Pol λ DL and wt Pol λ (Table 1).
Table 1.
Steady-state kinetic parameters for single nucleotide incorporation by Pol λ DL
| Enzymes | Primer terminus | xNTP | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | Discriminationa |
|---|---|---|---|---|---|---|
| Pol λ wt | ||||||
| dC | dATP | 0.22 ± 0.13 | 0.20 ± 0.025 | 0.9 ± 0.5 | ||
| dC | rATP | 50.6 ± 11.6 | 0.014 ± 0.004 | 2.8 × 10−4 ± 1 × 10−4 | 3200 | |
| rC | dATP | 0.19 ± 0.09 | 0.140 ± 0.039 | 0.74 ± 0.42 | 1.4 | |
| Pol λ DL | ||||||
| dC | dATP | 0.07 ± 0.01 | 0.075 ± 0.007 | 1.04 ± 0.19 | ||
| dC | rATP | 13 ± 3.7 | 0.069 ± 0.006 | 5.3 × 10−3 ± 1.6 × 10−3 | 200 | |
| rC | dATP | 0.07 ± 0.02 | 0.04 ± 0.008 | 0.63 ± 0.23 | 1.65 |
The values for the kinetic constants are an average of three to five independent determinations (mean ± SD).
aDiscrimination at the insertion step is defined as the ratio of catalytic efficiencies (kcat/Km)dATP/(kcat/Km)rATP and discrimination at the extension step is defined as the ratio of (kcat/Km)dC/(kcat/Km)rC for correct dATP insertion.
Structural analysis for rNTP incorporation and extension
Pre-catalytic complex for ribonucleotide incorporation (incorporation-ternary)
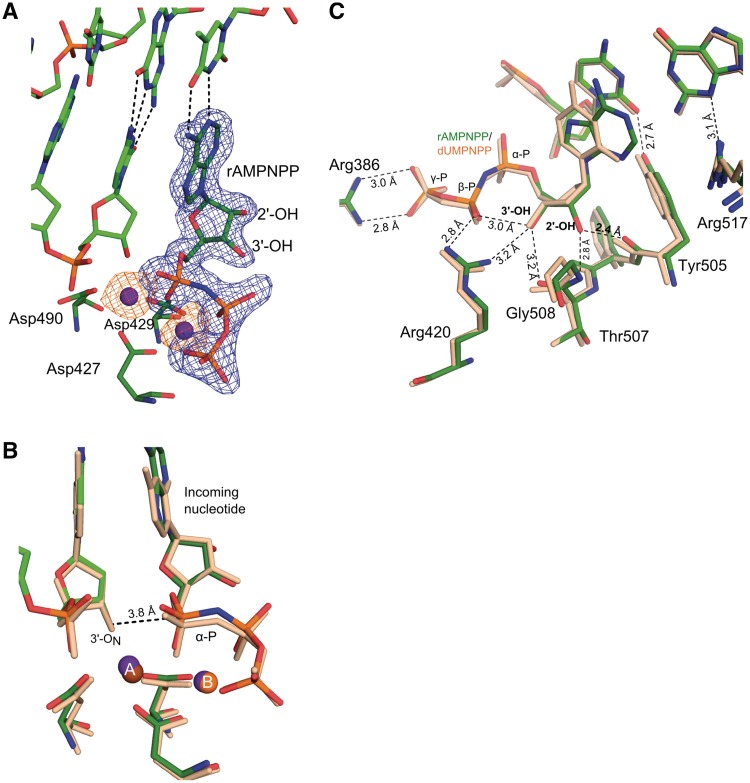
To understand how rNTPs are incorporated into DNA, a structure of the pre-catalytic complex of Pol λ DL with a one-nucleotide gap DNA substrate and an incoming non-hydrolyzable ribonucleotide analog, rAMPNPP, was obtained at 1.95 Å resolution (Figures 1A and 2, Supplementary Figure S1 and Table 2). This structure reveals rNTP binding in a catalytically competent geometry with no obvious distortion to the active site as compared to the ternary complex structure of wt Pol λ with an incoming dNTP [PDB ID code 2PFO (23), Supplementary Figure S1A]. Both protein and DNA are in an active conformation consistent with structures of other ternary complexes for Pol λ (16,18,23). The incoming rAMPNPP forms a canonical Watson–Crick base pair with the templating thymine base (Figure 2A). Two Mn2+ atoms are observed: one at the catalytic A and the other at the binding B site. Both metals complete their octahedral coordination via interaction with the ribonucleotide triphosphate, the catalytic aspartates 427, 429 and 490 and water molecules (Supplementary Figure S1B). The sugar puckers for the incoming rAMPNPP and dCMP at the −1 site (as defined in Figure 1) adopt the C3′-endo conformation, similar to the ternary complex with dNTP. The non-specific interactions between protein residues (Tyr505 and Arg517) and the minor groove of the DNA, which are involved in selecting the correct nucleotide, are also similar. The nucleophilic 3′-ON on the primer terminus is 3.8 Å away from the α-phosphate of the incoming nucleotide similar to that seen for the pre-catalytic complex of wt Pol λ (3.7 Å) which is consistent with a pre-catalytic state for an in-line nucleotidyl transfer reaction (Figure 2B). The 3′-OH of the incoming rAMPNPP lies within hydrogen bonding distance from the side chain of Arg420 (3.2 Å), the carbonyl of Thr507 (3.2 Å) and the β-phosphate oxygen (3.0 Å), and in addition Arg420 and Arg386 are within hydrogen bonding distances from the β- and γ-phosphate of the incoming rNTP, respectively, suggesting that these are stabilizing interactions. The 2′-OH is located at a surprisingly close distance of 2.4 Å from the carbonyl oxygen of Tyr505 and 2.8 Å from the backbone amide nitrogen of Gly508 (Figure 2C).
Figure 2.
Pre-catalytic complex for ribonucleotide incorporation (incorporation-ternary). (A) Active site of the ternary complex of Pol λ DL with bound rAMPNPP. Contoured at 3σ, the simulated annealing Fo-Fc omit density map (blue) and the anomalous difference density map (orange) for both Mn2+ ions (purple), are shown. (B) Superposition of Pol λ DL with wt Pol λ. The structure of the ternary complex of Pol λ DL:rAMPNPP (green) is superimposed with ternary wt Pol λ:dUMPNPP complex [PDB ID code 2PFO, wheat, (23)]. The distance between the nucleophile at the primer terminus, 3′-ON and the α-phosphate (α-P) is shown as dashed line. (C) Active site of Pol λ DL:rAMPNPP showing interactions for 2′-OH, 3′-OH, β- and γ-phosphates of the bound nucleotide in black dashed lines. Interactions of Tyr505 and Arg517 with bases at the −1 site are also shown in black dashed lines. α-, β- and γ-phosphates on the nucleotide are indicated as α-P, β-P and γ-P, respectively.
Table 2.
Summary of crystallographic data
| Parameters | Incorporation-ternarya | Incorporation-productb | Extension-ternarya | Extension-producta |
|---|---|---|---|---|
| Data collection | ||||
| Unit cell dimensions (Å) (a × b × c) | 55.98 × 63.66 × 139.97 | 55.91 × 64.13 × 139.18 | 55.78 × 63.66 × 141.47 | 55.29 × 62.00 × 141.47 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Resolution (Å) | 1.95 | 2.14 | 2.00 | 2.25 |
| No. of observations | 235 524 | 235 747 | 148 222 | 143 574 |
| Unique reflections | 37 285 | 27 507 | 32 824 | 23 003 |
| Rsym (%) (last shell) | 5.8 (35.7) | 10.7 (31.4) | 9.3 (29.7) | 6.9 (28.8) |
| I/σI (last shell) | 28.9 (3.7) | 20.4 (6.2) | 11.1 (2.9) | 23.7 (5.3) |
| Completeness (%) (last shell) | 99.8 (98.4) | 97.2 (90.1) | 95.5 (78.4) | 93.1 (69.7) |
| Refinement statistics | ||||
| Rcryst (%) | 20.8 | 19.5 | 18.8 | 20.6 |
| Rfree (%)c | 24.4 | 23.9 | 23.0 | 24.7 |
| Complexes in asymmetric unit | 1 | 1 | 1 | 1 |
| Mean B value (Å2) | 28.05 | 34.92 | 28.84 | 41.68 |
| Root mean square deviation from ideal values | ||||
| Bond Length (Å) | 0.010 | 0.006 | 0.011 | 0.008 |
| Bond angle (degrees) | 1.069 | 0.94 | 1.283 | 1.211 |
| Ramachandran statistics (22) | ||||
| Residues in favored regions | 95.5 | 96.4 | 99.4 | 96.8 |
| Residues in allowed regions | 100 | 100 | 100 | 100 |
aData collected at the SER-CAT beamline ID-22 beamline at the Advanced Photon Source, at Argonne National Laboratory.
bData collected on a Saturn92 CCD area detector mounted on a MicroMax-007HF (Rigaku Corporation) rotating anode generator equipped with Varimax HF mirrors.
cReference Rfree reflections were maintained (PDB ID code 3MGI).
Post-catalytic complex for ribonucleotide incorporation (incorporation-product)
A structure of the post-catalytic complex of Pol λ DL with a nicked DNA containing rAMP inserted opposite a templating thymine was obtained at a resolution of 2.15 Å (Figures 1B and 3, Supplementary Figure S2 and Table 2). This incorporation-product structure reveals significant differences in the active site, compared to that of a structure of wt Pol λ:dNMP product complex [PDB ID code 1XSP (18)]. With dNTP as the incoming nucleotide, completion of the nucleotidyl transfer reaction and advancement from ternary complex to the product complex involves no significant conformational changes of the protein or the DNA, other than stereochemical inversion of the α-phosphate [Figure 2b in (18)]. The post-catalytic complex of Pol λ DL with rNMP is similar to that of wt Pol λ with dNMP in that Watson–Crick base pairing is maintained, with no obvious distortion of the protein (PDB ID code 1XSP, Figure 3A and Supplementary Figure S2A). The catalytic metal (metal A) is not present in the structure since it dissociates after nucleotide incorporation. The same was observed in the structure of post-catalytic complex of wt Pol λ (18). Additionally, metal B is also not present in the structure. It appears that the pyrophosphate with the bound metal B is replaced by a sulfate ion that is present in high concentration in the crystallization condition. The sugar pucker of the inserted ribonucleotide is in the C3′-endo conformation, as is observed for an inserted dNMP (18). However, the deoxyribose sugar of the nucleotide at −1 site adopts a C2′-endo conformation, as opposed to a C3′-endo in the dNMP-incorporated structure (Figures 1B, 3A and 3B). Upon insertion of the rNMP, the 2′-OH is expelled from its position in the pre-catalytic complex and occupies a position normally occupied by the 3′-OH of the sugar, flipping the phosphate of the newly incorporated nucleotide by 4.2 Å into the minor groove and displacing the sugar at the −1 site by 3.5 Å (Figure 3B). The phosphate oxygens of the rAMP lie within hydrogen bonding distances to Lys472 (2.7 Å), the carboxyl of Asp429 (3.0 Å) and the hydroxyl of Tyr505 (2.7 Å) (Figure 3C). This distortion of the primer strand coincides with the movement of residues Tyr505 and Phe506 from their active conformation, as observed in the structure of wt Pol λ product complex with dNMP (Supplementary Figure S2B, PDB ID code 1XSP, wheat), to a position more similar to the inactive binary conformation (Supplementary Figure S2B, PDB ID code 1XSL, pink). In contrast, Arg514 and Arg517, which interact with the template strand, remain in their active conformations. There is no distortion of the template strand and the templating base remains properly positioned in the nascent base pair binding pocket.
Figure 3.
Post-catalytic complex for ribonucleotide incorporation (incorporation-product). (A) The structure of the post-catalytic complex of Pol λ DL with inserted rAMP is shown. A simulated annealing Fo-Fc omit density map (green, contoured at 3σ), is shown for the bases at 0 and −1 sites. (B) Superposition of post-catalytic complexes of Pol λ DL and wt Pol λ. The structure of the post-catalytic complex of Pol λ DL with inserted rAMP (blue) is shown in an overlay with post-catalytic complex of wt Pol λ [PDB ID code 1XSP, wheat, (18)]. Displacements in sugar-phosphate backbone are indicated by black arrows. (C) Possible interactions for α-phosphate oxygens with Tyr505, Lys472 and Asp429 are shown in dashed lines.
Pre-catalytic complex for extension of an rNMP-terminated primer (extension-ternary)
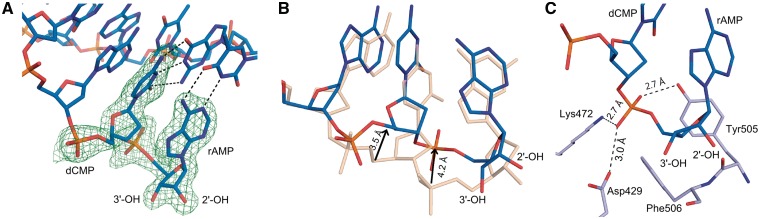
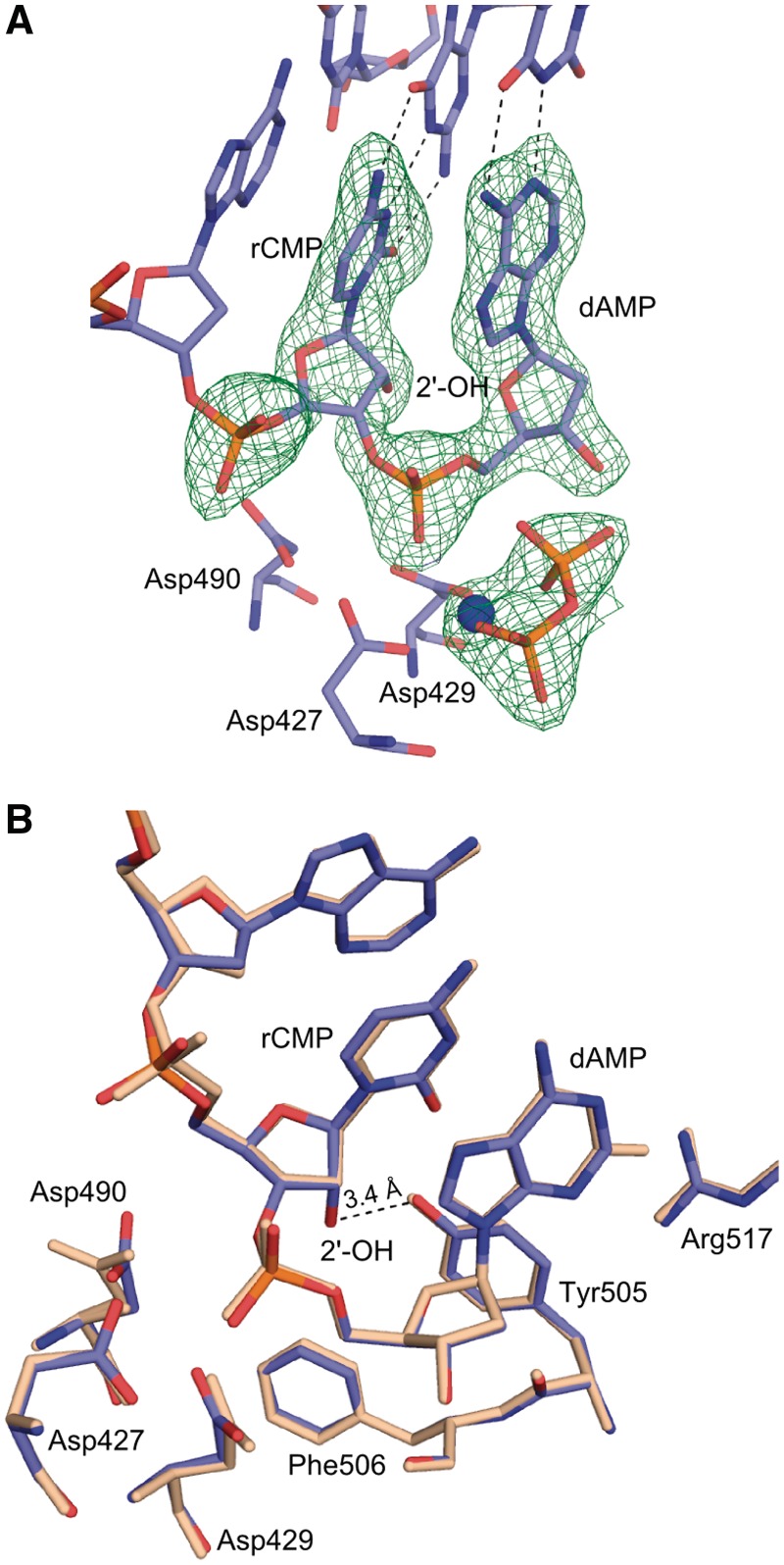
To gain insight into how the newly incorporated rNMP can function as a nucleophile for further polymerization, structures representing pre- and post-catalytic complexes of Pol λ DL with a rCMP-terminated primer were obtained. Crystals of the pre-catalytic ternary complex of Pol λ DL with rCMP at the primer terminus of a one-nucleotide gap DNA substrate and dAMPCPP at the incoming position opposite to a template thymidine diffracted to 2.0 Å resolution (Figures 1C and 4, Supplementary Figure S3 and Table 2). Once the newly incorporated ribonucleotide moves from the active site to the −1 position (Figure 1C), the distortion observed in the incorporation-product complex is no longer present. This pre-catalytic structure superimposes well with the pre-catalytic complex structure of wt Pol λ with a dNMP terminated primer (PDB ID code 2PFO, Supplementary Figure S3A). Both the incoming dAMPCPP and the primer-terminal rCMP form canonical Watson–Crick base pairs with the corresponding nucleotides in the template strand (Figure 4A). The sugar puckers for incoming dAMPCPP at 0 and rCMP at −1 sites are in C3′-endo conformation. The dAMPCPP is in a catalytically relevant position with a Mn2+ at the metal A site and a Mg2+ at the metal B site (Supplementary Figure S3B). There are no obvious changes in the position of the active site atoms. All relevant moieties are in the active conformation, including the position of the nucleophilic 3′-ON, located 3.4 Å away from the α-phosphate of the incoming dAMPCPP (Figure 4B). As seen in Figure 4B, the presence of the 2′-OH of the primer-terminal nucleotide is easily accommodated in the minor grove and is within hydrogen bonding distance to the hydroxyl of Tyr505. Clear density around the rCMP at the −1 site and the dAMPCPP at the 0 site indicate that the extension of rNMP-terminated primer will occur from the expected 3′-OH.
Figure 4.
Pre-catalytic complex for extension of an rNMP-terminated primer (extension-ternary). (A) The structure of the ternary complex of Pol λ DL:dAMPCPP (magenta) is shown. A simulated annealing Fo-Fc omit density map (contoured at 3σ), is shown in blue for the inserted ribonucleotide at the −1 position and incoming deoxyribonucleotide at the nascent base pair binding site and in green for the Mg2+ ion (Mg2+ ion shown in blue). Anomalous difference density map (orange, contoured at 3σ) is shown for Mn2+ ion (purple). (B) Superposition of Pol λ DL with wt Pol λ. The structure of Pol λ DL:dAMPCPP (magenta) is superimposed with ternary complex of wt Pol λ [PDB ID code 2PFO, gray (23)]. 2′-OH of the inserted rAMP is within hydrogen bonding distance to hydroxyl of Tyr505 (black dashed line). Also highlighted is the distance between the α-phosphate and the nucleophilic 3′-ON suggesting catalytically relevant position of the incoming nucleotide.
Post-catalytic complex for extension of an rNMP-terminated primer (extension-product)
A structure of the post-catalytic complex of Pol λ DL with nicked DNA at 2.25 Å resolution, containing a ribonucleotide-terminated primer strand extended by one dNMP, was captured by consecutive insertion of rCMP and dAMP at the −1 and 0 positions, respectively (Figures 1D and 5, Supplementary Figure S4 and Table 2). This extension-product structure reveals no major differences as compared to an extension-product structure of wt Pol λ with dNMPs at the corresponding positions (PDB ID code 1XSP, Figure 5B and Supplementary Figure S4). The sugar puckers for incorporated dAMP at 0 and rCMP at −1 sites are in C3′-endo conformation similar to the product complex for wt Pol λ (PDB ID code 1XSP). Watson–Crick base pairing is observed for both incorporated dAMP and rCMP with templating bases. Unlike with a newly incorporated rAMP in the nascent binding site, all protein and DNA residues remain in the active conformation.
Figure 5.
Post-catalytic complex for extension of an rNMP-terminated primer (extension-product). (A) The structure of the post-catalytic complex of Pol λ DL with incorporated dAMP (purple). A Mg2+ ion bound in the B site is shown in blue. Three catalytic aspartates Asp427, Asp429, Asp490 and the pyrophosphate (orange) are shown. A simulated annealing Fo-Fc omit density map, (green, contoured at 3σ), is shown for the incorporated rCMP and the dAMP at the −1 and 0 positions, respectively, as well as for Mg2+ ion and for the pyrophosphate. (B) Superposition of post-catalytic complexes of Pol λ DL (blue) and wt Pol λ (wheat) [PDB ID code 1XSP, (18)]. The 2′-OH of the rCMP at the −1 position is within hydrogen bonding distance to the hydroxyl of Tyr505 (shown).
DISCUSSION
Here, we describe four crystal structures of Pol λ DL that correspond to steps in the catalytic path for rNTP incorporation. The structure of the incorporation-ternary complex (Figures 1A and 2) reveals an unfavorable, repulsive interaction between the 2′-OH on the ribose of the incoming nucleotide and the backbone carbonyl of Tyr505. This suggests an energetically unstable binding state for the rNTP, which appears to be stabilized by additional interactions between the protein and the ribonucleotide that allow insertion. These interactions include hydrogen bonds of Arg420 and Arg386 with the β- and γ-phosphates of the rNTP, respectively (Figure 2C). In keeping with this hypothesis, the 200-fold lower catalytic efficiency for rNTP incorporation reflects a higher apparent Km for the ribonucleotide (Table 1). In contrast to the structure of the pre-catalytic incorporation complex, the structure of the post-catalytic incorporation complex (Figures 1B and 3), which represents the state after the new phosphodiester bond is formed but before DNA translocates, is highly distorted. A superposition of this structure with the structure of the post-catalytic incorporation-product complex of wt Pol λ with dNMP indicates distortion of the primer terminus (Figure 3B). The 2′-OH is expelled from its pre-catalytic position and occupies the same position as the 3′-OH in the dNMP containing complex (Figure 3B), the phosphate of the primer-terminal rNMP and the sugar of the preceding nucleotide (−1 site in Figure 1) are shifted into the minor groove (Figure 3B), and the hydroxyl of Tyr505 is not at hydrogen bonding distance from the base of the primer nucleotide at −1 site anymore (Figure 3C). We suggest that the distortion of the post-catalytic incorporation-product complex is a consequence of the energetically unstable binding state for the rNTP. Once the bond between the α- and β-phosphates of the rNTP is broken, the stabilizing interactions with the β- and γ-phosphates observed in the pre-catalytic incorporation-ternary complex structure are lost, such that the unstable binding state of the ribonucleotide can no longer be maintained.
A repulsive, short distance interaction, similar to the one in the Pol λ DL pre-catalytic incorporation-ternary complex, is observed between the 2′-OH and Tyr271 (Tyr505 in Pol λ) in the structure of a incorporation-ternary complex of Pol β with an incoming rNTP (15), which superimposes well with the structure of the corresponding complex of Pol λ DL (Supplementary Figure S1C). While there is no structure of the post-catalytic incorporation-product complex of Pol β for rNTP incorporation, the structure of the pre-catalytic incorporation-ternary complex together with quantum mechanical calculations based on the Pol β:rNTP ternary complex, also suggest that there is an energetic cost for accommodating the rNTP in the nascent base pair binding pocket (15). Despite the energetically unstable binding of the rNTP suggested by the ternary complexes for Pols λ and β, the post-catalytic incorporation-product complex for Pol λ (Figure 3) shows how a ribonucleotide can be stably incorporated by Pol λ, Pol β and perhaps by other family X polymerases.
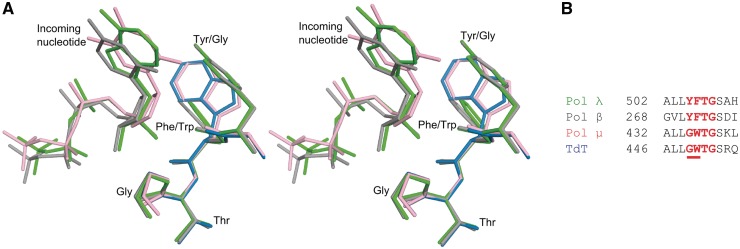
Based on the incorporation-ternary and incorporation-product structures (Figures 1A, 1B, 2 and 3), the energetic cost for accommodating the rNTP in the nucleotide binding pocket of Pol λ is largely a consequence of the steric clash between the 2′-OH of the rNTP and the backbone carbonyl of a tyrosine residue. Replacing Tyr505 in Pol λ with Gly or Ala and Tyr271 in Pol β with Phe or Ala reduces the sugar selectivity by about 10-fold for Pol λ (13) and up to 12-fold for Pol β (15,24,25). This suggests some role in rNTP discrimination for the side chain of the tyrosine residue, in addition to that of its backbone carbonyl. For example, one possibility is that replacement of the bulky tyrosine side chain with a residue containing a smaller side chain may affect the flexibility of the backbone so that the rNTP can now be more easily accommodated. It has been suggested that rNTP accommodation in the binding site may also be influenced by the interaction between the hydroxyl of Tyr505 and the primer terminus (15). The observations that the tyrosine side chain plays a role in rNTP discrimination is consistent with results indicating that TdT and Pol µ, which in contrast to Pols λ and β have a glycine residue instead of the tyrosine (Figure 6), discriminate against rNTPs 102- to 103-fold less efficiently than Pols λ and β. It is worth noting, that low sugar discrimination by TdT and Pol µ is also related to their significantly lower catalytic efficiency for dNTP incorporation relative to that of Pols λ and β (10).
Figure 6.
Comparison of the active site regions for X family polymerases. (A) Stereoview of the superposition of ternary pre-catalytic structures of Pol λ DL:rAMPNPP (green), Pol β:rCTP [PDB ID code 3RH4, gray (15)], Pol µ:ddTTP [PDB ID code 2IHM, pink (26)] and apo TdT [PDB ID code 1JMS, blue (27)]. (B) Sequence alignment for Pol λ, Pol β, Pol µ and TdT in the residue range used for superposition.
It is not clear if, and how, the distortion of the post-catalytic insertion complex affects translocation of the newly inserted rNMP from the active site to the −1 position. For example, it could promote translocation or destabilize the polymerase–DNA interaction causing the enzyme to dissociate. In fact, even the translocation of a dNMP-terminated primer bound by Pol λ is not well understood. This is a complex problem because Pol λ, like other family X enzymes, has a unique mode of DNA binding during short gap-filling synthesis. It has at least two DNA binding sites. Its polymerase domain binds the primer-template, while its 8 kDa domain interacts with the 5′-end of the gap. Thus, even if the polymerase domain dissociates from the primer-template, the enzyme may remain tethered to the DNA by its 8 kDa domain. This allows the polymerase domain to rebind the template-primer. The structures of the pre- and post-catalytic extension complexes (extension-ternary and extension-product, respectively) of Pol λ DL (Figures 1C, 1D, 4 and 5) suggest that once a newly incorporated ribonucleotide is rebound at the −1 position (Figure 1C), the distortion of the primer terminus observed in the incorporation-product complex (Figures 1B and 3) is no longer present, allowing for further extension. The structures of the extension-ternary and the extension-product complexes overlay well with the corresponding structures of wt Pol λ with a dNMP-terminated primer (Figures 4B and 5B) further indicating that the rNMP at the primer terminus is accommodated without any distortion to the DNA or the protein. Consistent with the structural data, the catalytic efficiency for incorporation of dATP onto a rNMP-terminated primer is similar to that for incorporation onto a dNMP-terminated primer (Table 1). In agreement with the Pol λ results, kinetic studies reported for wt Pol β show that the catalytic efficiencies for dNTP incorporation onto a rNMP-terminated and dNMP-terminated primer are also similar (14). Thus, once the ribonucleotide is incorporated, the polymerase appears to have no problem extending a primer-terminal rNMP. This indicates that Pol λ, and Pol β (15), discriminate against rNTPs at the incorporation step.
As suggested in our earlier work (16), the conformational transition from the inactive to the active state captured in the structures (Figures 2 and 4) may follow a different path for Pol λ DL relative to that for wt Pol λ. Nevertheless, the fact that the structures of pre-catalytic complexes for Pol λ DL incorporating a ribonucleotide (incorporation-ternary, Figure 2B) or a deoxyribonucleotide [Figure 5A in (16)] overlay well with that for wt Pol λ incorporating a deoxyribonucleotide, supports the idea that Pol λ DL is a good structural model for the wild-type polymerase.
The described structures are the first to present a complete catalytic cycle of rNTP incorporation and extension by a DNA polymerase, providing a foundation for understanding stable ribonucleotide incorporation into DNA. The structure of the pre-catalytic incorporation-ternary complex (Figures 1A and 2) with a non-hydrolyzable analog of rATP, described here, contains all atoms required at the active site in a configuration consistent with catalysis. Moreover, the structure of the incorporation-product complex (Figures 1B and 3) highlights the consequences of the energetically unfavorable interaction in the structure of the incorporation-ternary complex providing additional insights into rNTP discrimination by Pol λ. The catalytic cycle for stable rNTP incorporation by Pol λ may be particularly relevant for repair of double-strand DNA breaks by NHEJ which is the major pathway for DSBR in non-proliferating cells. In non-proliferating cells, the concentration of dNTPs relative to rNTPs is even lower than in cycling cells, further increasing the likelihood of rNTP incorporation. In such situations, incorporation of an rNTP may be needed for the cell to escape the consequences of an unrepaired double-strand DNA break. The proficiency of family X polymerases to incorporate rNTPs may be particularly beneficial in such cases.
We believe that these new structural insights into the catalytic cycle for rNTP incorporation by a DNA polymerase are an important contribution to studies on the potential functions and biological consequences of rNTP incorporation into the genome, an evolving chapter in research on genome stability.
ACCESSION NUMBERS
Data deposition: the structure factors have been deposited in the PDB, www.pdb.org.
Crystal structure of pre-catalytic ternary complex of polymerase λ variant with a rATP analog opposite a templating T (incorporation-ternary) has been assigned the RCSB ID code rcsb069054 and PDBID code 3UPQ.
Crystal structure of post-catalytic product complex of polymerase λ variant with rAMP at the primer terminus (incorporation-product) has been assigned the RCSB ID code rcsb069064 and PDB ID code 3UQ0.
Crystal structure of pre-catalytic ternary complex of polymerase λ variant with a dATP analog opposite a templating T and a rCMP at the primer terminus (extension-ternary) has been assigned the RCSB ID code rcsb069065 and PDBID code 3UQ1.
Crystal structure of post-catalytic product complex of polymerase λ variant with a rAMP inserted opposite a templating G and dAMP inserted opposite a templating T at the primer terminus (extension-product) has been assigned the RCSB ID code rcsb069066 and PDB ID code 3UQ2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4, Supplementary Methods and Supplementary References [15,16,18,23].
FUNDING
The Division of Intramural Research of the National Institutes of Health, National Institute of Environmental Health Sciences [Project Z01 ES065070 to T.A.K. and Project Z01 ES102645 to L.C.P. in parts]. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. Funding for open access charge: Division of Intramural Research of the National Institutes of Health, National Institute of Environmental Health Sciences Project [Z01 ES065070 to T.A.K.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs W.C. Copeland and W.B. Beard for critical reading of this article.
REFERENCES
- 1.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirouac KN, Suo Z, Ling H. Structural mechanism of ribonucleotide discrimination by a Y-Family DNA polymerase. J. Mol. Biol. 2011;407:382–390. doi: 10.1016/j.jmb.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 3.Astatke M, Ng K, Grindley NDF, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl Acad. Sci. USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol α DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 5.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundström E-B, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundström E-B, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 8.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl Acad. Sci. USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulé J-B, Rougeon F, Papanicolaou C. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 10.Nick McElhinny SA, Ramsden DA. Polymerase Mu is a DNA-directed DNA/RNA polymerase. Mol. Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz JF, Juárez R, García-Díaz M, Terrados G, Picher AJ, González-Barrera S, Fernández de Henestrosa AR, Blanco L. Lack of sugar discrimination by human Pol µ requires a single glycine residue. Nucleic Acids Res. 2003;31:4441–4449. doi: 10.1093/nar/gkg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 13.Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. A novel mechanism of sugar selection utilized by a human X-Family DNA polymerase. J. Mol. Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanaugh NA, Beard WA, Wilson SH. DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavanaugh NA, Beard WA, Batra VK, Perera L, Pedersen LG, Wilson SH. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J. Biol. Chem. 2011;286:31650–31660. doi: 10.1074/jbc.M111.253401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bebenek K, Garcia-Diaz M, Zhou R-Z, Povirk LF, Kunkel TA. Loop 1 modulates the fidelity of DNA polymerase λ. Nucleic Acids Res. 2010;38:5419–5431. doi: 10.1093/nar/gkq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bebenek K, Pedersen LC, Kunkel TA. Replication infidelity via a mismatch with Watson–Crick geometry. Proc. Natl Acad. Sci. USA. 2011;108:1862–1867. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol λ catalytic cycle. Nat. Struct. Mol. Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CWJ, Sweet RM, editors. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [Google Scholar]
- 20.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Role of the catalytic metal during polymerization by DNA polymerase lambda. DNA Repair. 2007;6:1333–1340. doi: 10.1016/j.dnarep.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 26.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase μ. Nat. Struct. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 27.Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.