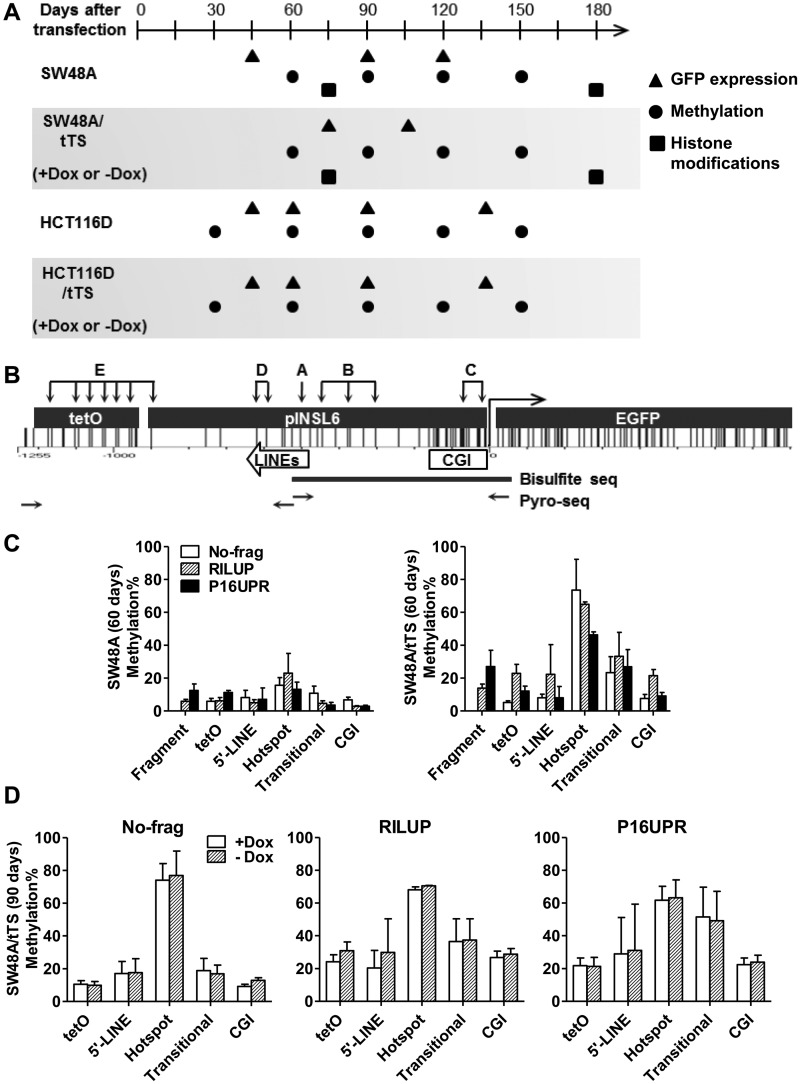
Figure 2.
De novo DNA methylation in the INSL6 promoter. (A) A graph showing the time points to collect long-term cultured cells after stable transfection. The coordinate is the time-scale (days) after constructs were transfected into Flp-in (SW48A and HCT116D) or Flp-in/tTS (SW48A/tTS and HCT116D/tTS) cells. Stable clones were cultured in media supplied with doxycycline (Dox) until 60 days (SW48A/tTS) or 30 days (HCT116D/tTS) before they were split and cultured separately in media with Dox (+Dox) and without Dox (−Dox). Symbols indicate the time to examine GFP expression (triangles), DNA methylation (circles) and histone modifications (squares). (B) Graphical distribution of CG sites in tetO-pINSL6-EGFP. The subcloned 940-bp pINSL6 consists of part of the CGI and two short LINE elements. Primers (horizontal arrows) for the first step of amplification of bisulfite-converted DNA include the tetO sequence or 5′-end of EGFP to distinguish it from the endogenous pINSL6. The capitals and groups of vertical arrows indicate the target sites for pyrosequencing (A, hotspot; B, Transitional; C, CGI; D, 5’-LINE; E, tetO). Assays for fragments are not shown here. Thick line, the amplified region for bisulfite cloning/sequencing. (C) Regional methylation of transgenes (No-frag, RILUP and P16UPR) in SW48A and SW48A/tTS (60 days). (D) Comparison of regional methylation levels of transgenes (No-frag, RILUP and P16UPR) in SW48A/tTS (90 days) under +Dox and −Dox.