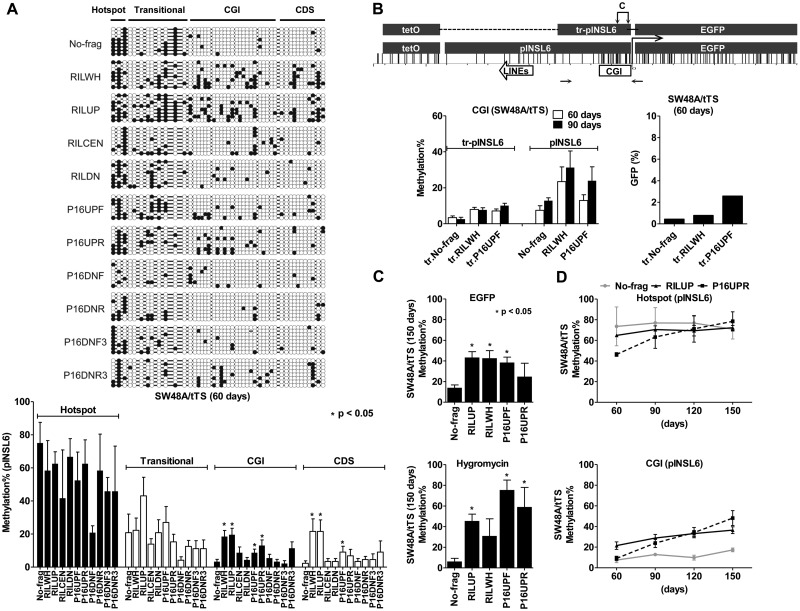
Figure 3.
Methylation spreading from the seeding sites to the adjacent regions. (A) Methylation patterns of the INSL6 promoter in 11 transgenes (SW48A/tTS, 60 days). Bisulfite-cloning/sequenced CGs are displayed in circles (closed, methylated; open, unmethylated); the average levels, calculated in respect to four regions (hotspot, transitional, CGI and CDS), are shown in the bar graph (mean ± SEM). *Statistically significant difference in comparison to No-frag (t-test, P < 0.05). (B) Removal of the seeding sites impaired CGI methylation in SW48A/tTS. The graph shows the distribution of CG sites in the truncated INSL6 promoter (tr-pINSL6), which retains the CGI and the tetO cassette. Horizontal arrows, the amplicon used for pyrosequencing (C, assays for CGI methylation). Pyrosequencing was performed for transgenes (No-frag, RILWH and P16UPF) in their truncated and complete forms (60 and 90 days). The repression of the truncated transgenes at 60 days was measured by GFP expression (flow cytometry). (C) Methylation extended to regions of EGFP and hygromycin sequences in SW48A/tTS (150 days). Pyrosequencing was used to examine 5 transgenes (No-frag, RILUP, RILWH, P16UPF and P16UPR). *Significantly methylated transgenes in comparison to No-frag (P < 0.05). (D) Gradual accumulation of transgene methylation in SW48A/tTS over time (60, 90, 120 and 150 days). Please refer to Supplementary Figure 5B for the cases of HCT116D and HCT116D/tTS.