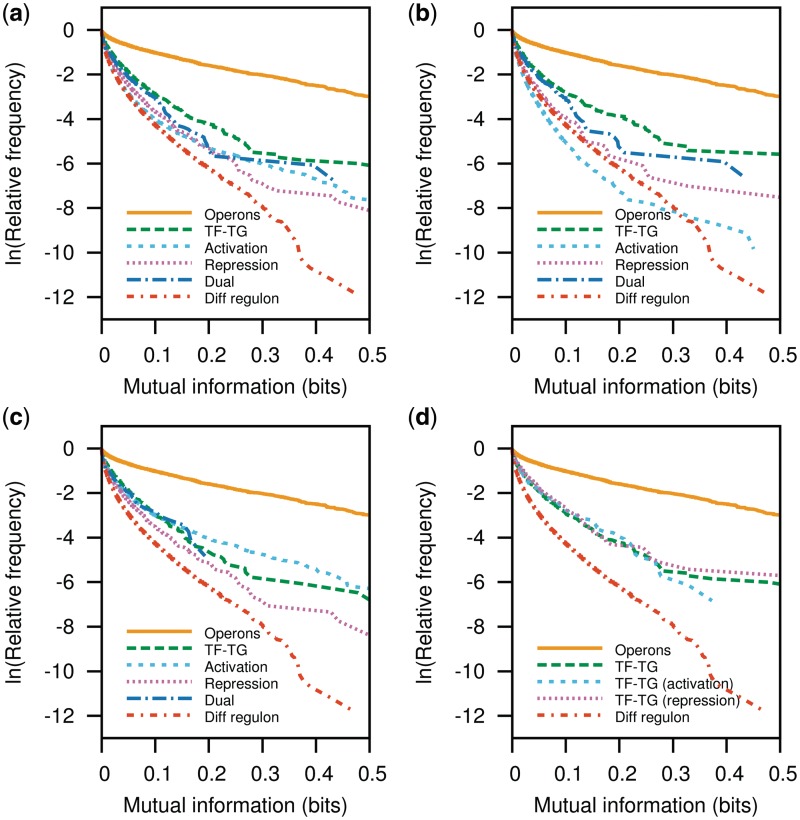
Figure 4.
P-cubic comparison of regulon subsets. Herein, we compared the P-cubic of positively co-regulated genes (activation), negatively co-regulated genes (repression) and the relationship between TFs and their TGs (TF–TG). (a) Overall, the most stable functional association corresponds to that between TFs and their TGs. Co-activated genes and dually regulated genes are next in stability with the lowest stability presented among co-repressed gene pairs. The relationships change when we analyse the subset of regulatory interactions by global regulators in (b). Although the TF–TG relationship shows the highest stability again, co-activated genes show a lower conservation than genes in different regulons, whereas dually co-regulated genes show the highest conservation. In the analyses involving local regulators (c), co-activated genes show more conservation than the TF–TG relationship. In agreement with previous results about the conservation of the TF–TG relationship among evolutionarily close Enterobacteria (38), we found that a higher proportion of repressor–TG relationships attain a higher mutual information than activator–TG relationships. However, even the most conserved interactions brought about by TFs remain close to those among overall TU borders shown in Figure 3b.