Figure 2.
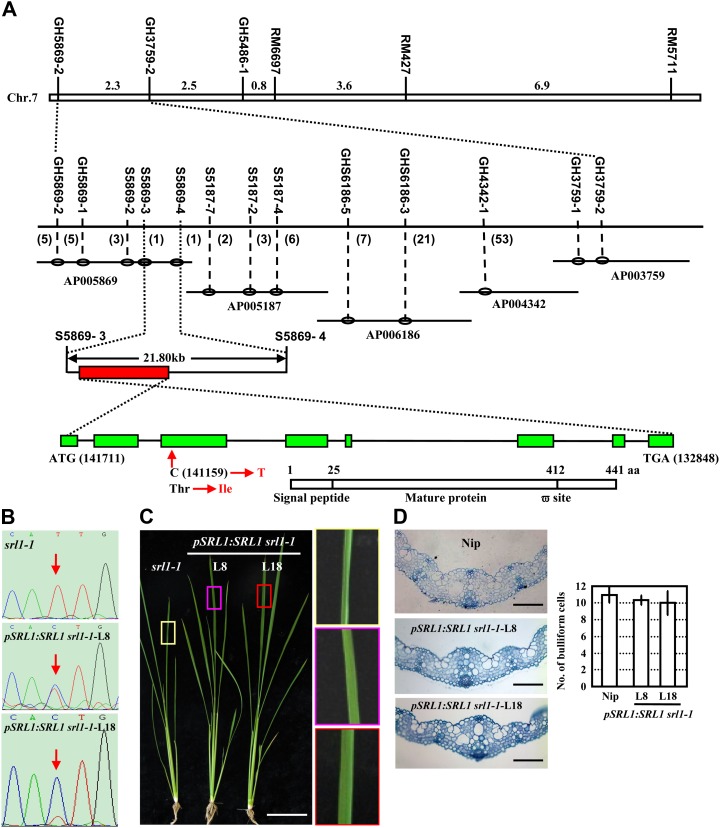
Map-based cloning of SRL1, which encodes a putative GAP. A, SRL1 was primarily mapped to rice chromosome 7 between STS markers GH5869-2 and GH3759-2 and then fine mapped to a 21.8-kb region between SSR markers S5869-3 and S5869-4 by a large F2 mapping population. Further amplification, sequencing, and comparison of the relevant DNA fragments indicated that a single base substitution (C to T) in the third exon of the gene Os07g01240 in srl1-1 allele leading to the change of amino acid residue from Thr to Ile. Genomic organization of SRL1 (green boxes represent exons, and black lines represent introns) is shown, including the translational start (ATG) and stop (TAA) positions of the predicted coding sequence. The position and nucleotide change in the srl1-1 mutant is also shown. Structural analysis of SRL1, a putative GAP, indicated the presence of an N-terminal signal peptide from 1 to 25 amino acids and a potential C-terminal GPI-modification site at 412 amino acids (ω-site). B, Sequencing analysis of the SRL1 transcripts in two transgenic lines (L8, L18) of srl1-1 with complemented expression of SRL1 showed the double peak at the mutation site (red arrows), indicating the presence of normal transcripts of SRL1. C and d, Morphology (C, 40-d-old plants, bar = 10 cm) and cross sections (D, third leaf blades of 14-d-old seedlings, bars = 100 μm) of two transgenic lines of srl1-1 with complemented expression of SRL1 (L8, L18). To highlight the leaf shape, leaf regions (boxes) are enlarged (right section of C). Rescued expression of SRL1 resulted in normal leaves and bulliform cell numbers. Statistical analysis by two-tailed Student’s t test shows no significant difference (right section of D). Error bar represents sd (n > 10).