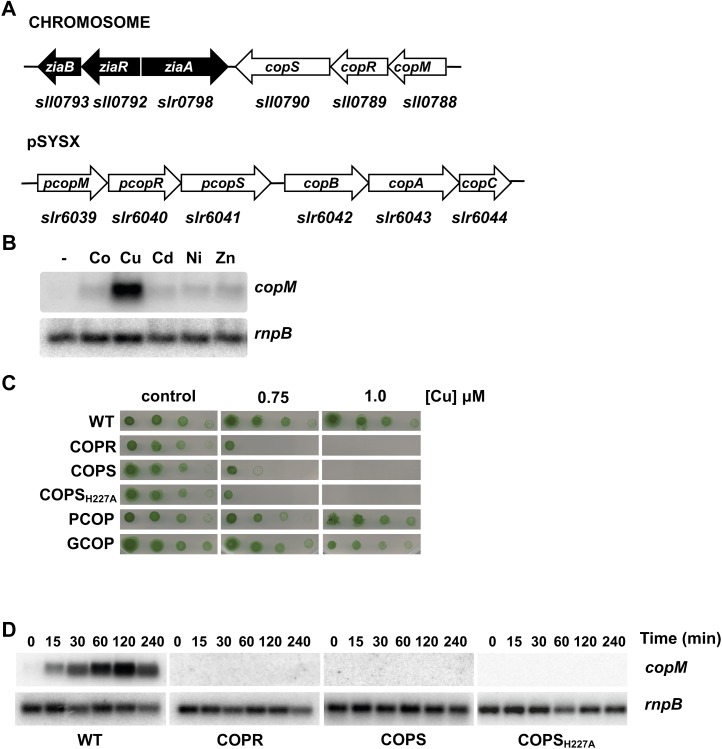
Figure 1.
CopRS is involved in copper resistance. A, Schematic representation of copMRS and pcopMRS-copBAC genomic regions. B, Northern-blot analysis of the expression of copM. Total RNA was isolated from wild-type cells grown in BG11C-Cu medium and exposed for 90 min to 3 μm of the indicated metal ions. Control cells were not exposed to added metals (−).The filter was hybridized with a copM probe and subsequently stripped and rehybridized with an rnpB probe as a control. C, Phenotypic characterization of mutants in copRS. Tolerance of wild-type, COPR, COPS, COPSH227A, PCOP, and GCOP strains to copper was examined. Ten-fold serial dilutions of a suspension of 1 μg chlorophyll mL−1 cells were spotted onto BG11C-Cu supplemented with the indicated copper concentrations. Plates were photographed after 5 d of growth. D, Loss of copM induction in COPR, COPS, and COPSH227A strains. Total RNA was isolated from wild-type, COPR, COPS, and COPSH227A strains grown in BG11C-Cu medium after addition of 3 μm of copper. Samples were taken at the indicated times. The filter was hybridized with a copM probe and subsequently stripped and rehybridized with an rnpB probe as a control. [See online article for color version of this figure.]