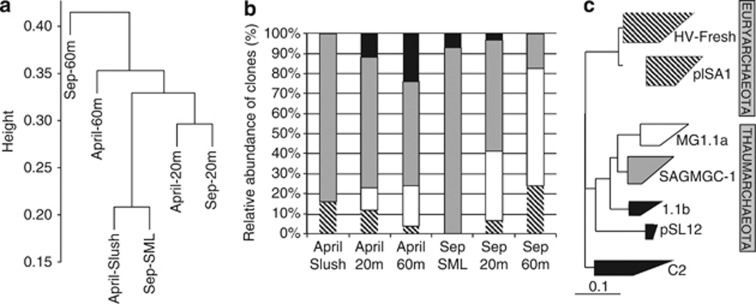
Figure 4.
Different analyses carried out with sequence data from the six 16S rRNA clone libraries of Lake Redon in April and September. (a) Hierarchical clustering analysis (UPGMA algorithm with Jackknife supporting values, 100 replicates). Distances between clusters expressed in UniFrac units (distance 0 means identical environments, distance 1 means two environments containing mutually exclusive lineages). (b) Archaeal community structure (relative abundances). Thaumarchaeota Group 1.1a (white label) and SAGMAGC-1 (gray label) are the dominant groups. Other minor thaumarchaeotal/crenarchaeotal groups detected are shown together (black label), as well as the euryarchaeotal groups (shaded label). (c) Collapsed maximum parsimony tree showing the phylogenetic position of the archaeal groups detected. See more details in Figure 5. Scale bar, 10% estimated divergence.