Abstract
Acid production from rhamnose is a characteristic phenotype of Listeria monocytogenes. We report the identification of the rhamnose transport and utilization operon located at lmo2846 to lmo2851, including the rhamnose-dependent promoter Prha. Expression of reporter genes under control of Prha on a single copy integration vector demonstrated its suitability for inducible gene expression in L. monocytogenes. Transcription initiation from Prha is dose dependent, and a concentration as low as 100 µM rhamnose was found sufficient for induction. Moreover, Prha is subject to glucose catabolite repression, which provides additional options for strict control of expression. Infection of human THP1 macrophages revealed that Prha is repressed in intracellular L. monocytogenes, which is explained by the absence of rhamnose in the cytosol and possible interference by catabolite repression. The Prha promoter provides a novel and useful tool for triggering gene expression in extracellular L. monocytogenes, whereas intracellular conditions prevent transcription from this promoter.
Introduction
Listeria monocytogenes is a non-sporulating Gram-positive rod, and the causative agent of human Listeriosis, a severe infection transmitted via contaminated food. The bacterium serves as a model organism for studies in cellular microbiology, bacterial pathogenicity and virulence, bacteriophage biology, and food safety [1], [2], [3], [4].
L. monocytogenes is able to produce acid from rhamnose, whereas L. innocua, L. welshimeri, and L. grayi show variable rhamnose utilization, and strains of L. ivanovii, L. seeligeri, and L. marthii are negative [5], [6]. Rhamnose is a naturally occurring L-6-deoxy hexose. It is present as a substituent of pectin in plant cell walls where it is periodically attached via α-1, 2-glycosylic linkages to galacturonic acid. In L. monocytogenes serovar 1/2 strains, rhamnose is not only used as a carbon source, it is also found as a decoration of the cell wall teichoic acids, and required for adsorption of A118 like bacteriophages [7].
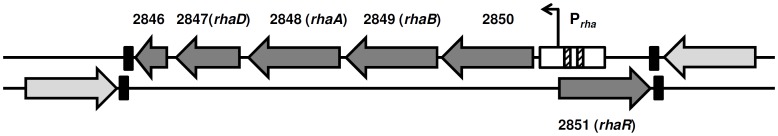
In Listeria, metabolism of rhamnose results in production of 1,2-propanediol, which is further oxidized to propionate [8]. In silico analysis indicated that rhamnose catabolism in both L. monocytogenes and L. innocua correlates with the presence of a set of genes clustered in a rhamnose operon (lmo2846-lmo2850 and lin2978-lin2982, respectively) [9], which encodes an ABC-transporter, a rhamnulokinase, rhamnose isomerase, rhamnulose-1-phosphate aldolase, and a rhamnose epimerase. An AraC-type DNA binding transcriptional regulator (lmo2851 and lin2983, respectively) is located immediately adjacent (Fig. 1). While the function of rhamnose operon genes and products has not been experimentally confirmed in Listeria, the specific uptake of rhamnose and the molecular biology and transcriptional regulation of these genes and their products has been thoroughly studied in E. coli. Remarkably, the structure and composition of the E. coli rha operon differs considerably from L. monocytogenes. While a rhamnose epimerase is not present, E. coli encodes two AraC-type regulatory proteins, RhaS and RhaR, to control rhaBAD expression [10], [11], [12], [13], [14], [15].
Figure 1. The L. monocytogenes rhamnose utilization operon.
Dark grey: open reading frames lmo2846-lmo2851 located on both DNA strands [9] and their functional assignments; white: Prha promoter, putative -10 and -35 regions are shaded; black: transcription terminators.
Gene products involved in carbohydrate transport and metabolism are often highly conserved yet specific for different prokaryotes. While E. coli encodes several carbohydrate transporters, e.g., the rha-, ara- and lac operons, facilitating rhamnose, arabinose and lactose uptake, respectively, B. subtilis features the xyl operon for xylose utilization. Gene expression and synthesis of carbohydrate transporters and accessory proteins is strictly regulated, and often depends on glucose-mediated catabolite repression [16], [17]. In this case, gene expression (i.e., repressor inactivation) only occurs when glucose is either not present or removed from the growth medium and replaced by the respective carbohydrate. Such tight control of promoter activity by repressor proteins can be elegantly harnessed for the design of inducible gene expression systems and vectors [18], [19].
With respect to L. monocytogenes, inducible gene expression is a highly desirable tool to study not only its pathogenicity and virulence, but also to better understand the environmental growth properties and responses of this opportunistic pathogen. Until now, inducible gene expression in this organisms has been based upon use of the lacI repressor and isopropyl-β-D-1-thiogalactopyranoside (IPTG) as an inducer [20], [21]. A disadvantage of this system is its poor stringency and often high background expression when a strong promoter is used. In fact, poor tune-ability and high level of read-through transcription can make it difficult or impossible to obtain quantitative data, or to clone or express toxic genes (L. Fieseler and M. J. Loessner, unpublished data). In contrast, positively-regulated gene expression (e.g., by an arabinose or rhamnose-dependent DNA binding AraC-type repressor) responds much slower following induction, while transcription repression is much tighter compared to negatively-regulated systems such as the lac operon [22].
Therefore, the aim of this study was to develop a tightly regulated, rhamnose-inducible gene expression system for Listeria, based on the Prha promoter. To monitor transcription activity, reporter genes encoding green fluorescent protein and drug resistance were placed under control of Prha on a single-copy integration vector. Employing both in vitro growth and intracellular infection models, we demonstrate that Prha enables quantitative expression of target genes, which can be modulated by the presence and concentration of rhamnose.
Results
Identification of the L. monocytogenes Prha Promoter
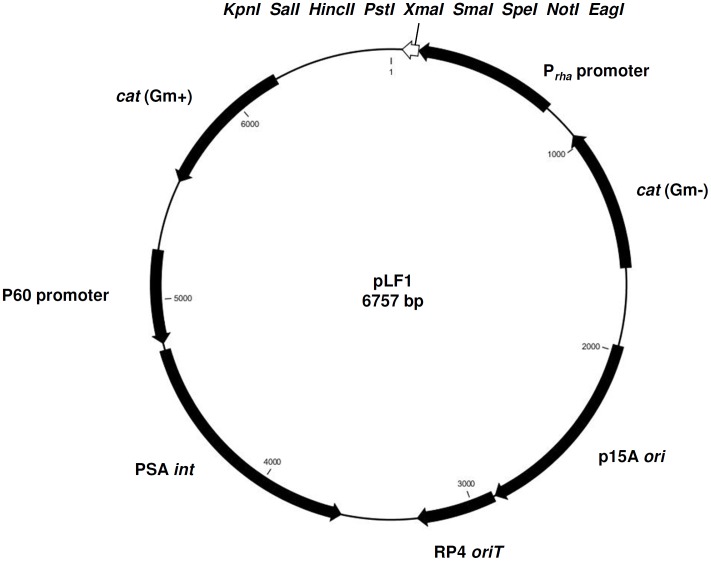
Putative -10 and -35 regions of a promoter designated as Prha were identified in silico immediately upstream of lmo2850, the first gene of a putative rhamnose utilization operon in L. monocytogenes (Fig. 1) [9]. In order to be able to test Prha for inducible gene expression, a 671 bp DNA fragment was amplified from the region upstream of the Shine-Dalgarno sequence of lmo2850 (nucleotide positions 2939946–2940597), and inserted into plasmid pPL2 to yield pLF1 (Fig. 2). Downstream of Prha, pLF1 still features the very useful multiple cloning site from pPL2, i.e., unique EagI, NotI, SpeI, SmaI, XmaI, PstI, HincII, SalI, and KpnI sites available for insertions and cloning.
Figure 2. Plasmid pLF1 for rhamnose-dependent gene expression in Listeria.
The Prha promoter sequence was inserted into the multiple cloning site of the single copy integration shuttle vector pPL2 [35]. The indicated restriction sites EagI, NotI, SpeI, SmaI, XmaI, PstI, HincII, SalI, and KpnI remain available for downstream insertions.
Transcription from Prha in Listeria is Rhamnose-dependent
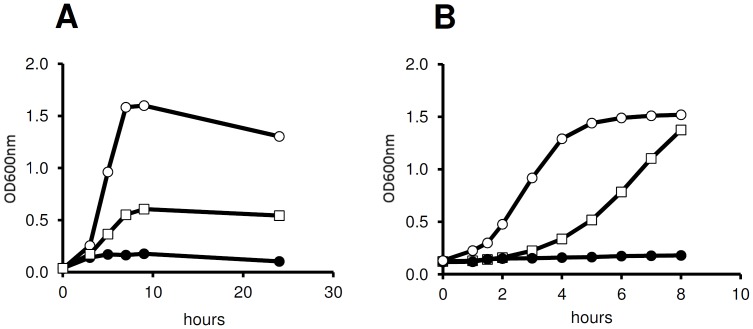
L. monocytogenes is able to utilize rhamnose (50 mM) as a carbon source during growth (Fig. 3A). Cells entered log phase after approximately 2 h, entered stationary phase after approximately 7 h, featuring an optical density (OD600 nm) of approximately 0.6. In the positive control (50 mM glucose), the culture reached a higher maximum OD600 nm of 1.6 after 7 h incubation, while only poor growth was observed in LB broth without additional carbon source (negative control).
Figure 3. Rhamnose dependent growth of L. monocytogenes in batch culture.
Panel A: Growth of L. monocytogenes 10403S in LB broth supplemented with glucose (50 mM, open circle) and rhamnose (50 mM, open squares), respectively. When no carbon source was added (closed circles), L. monocytogenes showed only poor growth. Panel B: Growth of L. monocytogenes LF002 (ermC under control of Prha) in medium supplemented with 7.5 µg/ml erythromycin. LF002 cells pre-induced with rhamnose (open squares) showed a slight delay in growth response, whereas non-induced cells (closed circles) did not multiply. L. monocytogenes strain LF003 (constitutive ermC expression) was used as positive control (open circles).
To determine whether expression from Prha is actually rhamnose-dependent, ermC was used as a reporter. L. monocytogenes LF002 was incubated in the presence of 10 mM rhamnose, before addition of 7.5 µg/ml erythromycin to the culture. While non-induced bacteria did not multiply further, rhamnose-induced cells continued growth and reached an OD600 nm of 1.4 after 8 h of incubation (Fig. 3B). The positive control (L. monocytogenes LF003 with constitutive expression of ermC) featured a shorter lag phase and faster onset of growth, and reached the stationary phase after 6 h of incubation (OD600 nm of 1.5).
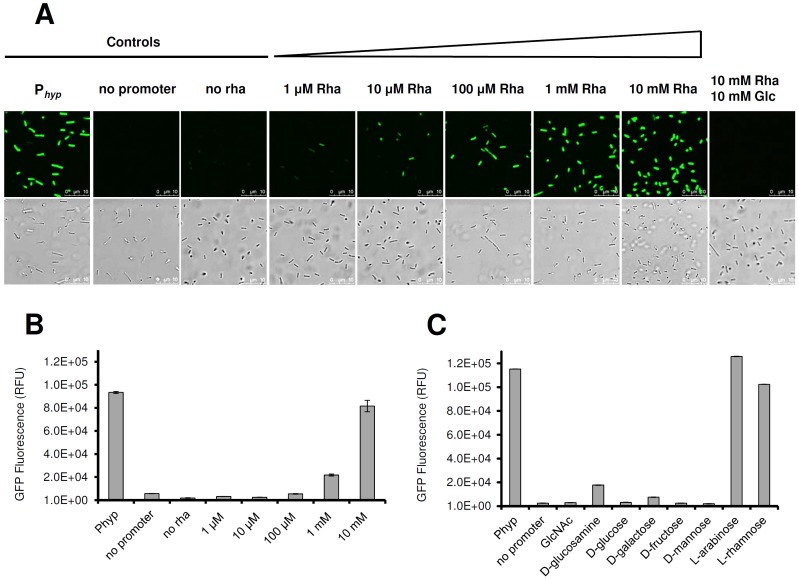
As an alternative to drug selection, the gfp gene was employed to measure expression from Prha in strain LF001, by monitoring green fluorescence following dose-dependent rhamnose induction. As negative control, LF004 (no promoter) was used, and LF005 (constitutive gfp expression from Phyp) served as positive control. We show that a concentration of 100 µM rhamnose was sufficient to quantitatively induce Prha-dependent transcription in a majority of cells (Fig. 4, panels A and B). Increasing rhamnose concentration of up to 10 mM resulted in stronger fluorescence, indicating that induction is dose-dependent. Even higher rhamnose concentrations did not result in a further increase of fluorescence, indicating that a plateau was reached. Interestingly, the presence of 10 mM glucose, N-acetylglucosamine, glucosamine, galactose, mannose, or fructose in addition to 10 mM rhamnose completely repressed Prha activity, demonstrating that transcription of the lmo2846-lmo2850rhamnose operon is subject to a strict catabolite repression. Interestingly, arabinose was the only sugar tested that did not affect Prha activity (Fig. 4C).
Figure 4. Rhamnose-inducible expression of GFP.
Panels A and B: Dose-dependent response of Prha-controlled gfp expression in L. monocytogenes LF001 after 16 h of induction using rhamnose concentrations as indicated. Panel A, top row: fluorescence microscopy; bottom row, phase contrast microscopy. Positive and negative controls, and the effect of catabolite repression by addition of equimolar amounts of glucose and rhamnose are indicated. rha: rhamnose, glc: glucose. Panel B: quantitation of relative fluorescence (RFU) of GFP in rhamnose-induced bacteria. Positive (Phyp) and negative (no promoter) controls are indicated on the left. Addition of 100 µM rhamnose increased fluorescence significantly (p<0.001). Panel C: quantitative catabolite repression of Prha-dependent gfp expression in the presence of rhamnose together with a second carbohydrate (indicated on the x-axis), at equimolar concentration (10 mM). Positive (Phyp) and negative (no promoter) controls are indicated on the right.
Prha is Repressed in Intracellular L. monocytogenes
It was interesting to determine if rhamnose would also be useful for regulating gene expression in intracellular L. monocytogenes during infection and the intracellular state. In the human intestine, rhamnose is not digested and regarded as a soluble dietary fiber material. While rhamnose as an osmolyte may affect cultured mammalian cells at higher concentrations, the amounts required for activation of Prha are quite low. We did not observe any negative effect on viability when THP1 macrophages [23] were exposed to 100 mM rhamnose over a period of 40 h. Cells maintained their shape and remained attached to the bottom of the cell culture flask, indistinguishable from controls without rhamnose (data not shown).
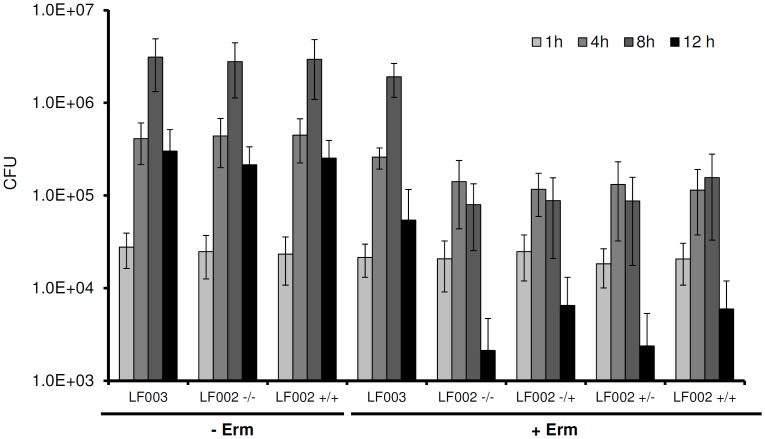
To determine transcriptional activity from Prha in L. monocytogenes during infection, strain LF002 that features ermC under control of Prha was used. We found that neither induced nor non-induced bacteria were able to multiply intracellularly in infected THP1 macrophages in presence of erythromycin (Fig. 5). Moreover, addition of rhamnose to infected macrophages did not result in ermC expression, i.e., the intracellular bacteria were unable to grow and multiply in the presence of the antibiotic. ANOVA analyses showed that the corresponding intracellular cell counts did not differ significantly from each other. In contrast, strain LF003 with constitutive expression of ermC was not affected, and significantly increased by 1.8 logs after an 8 h infection period (p<0.0001). In the absence of the drug, all strains showed identical growth responses regardless if exposed to rhamnose or not (Fig. 5).
Figure 5. Intracellular multiplication of L. monocytogenes LF002 (ermC under control of Prha) in human THP1 macrophages, in the presence and absence of rhamnose and erythromycin (see Material and Methods for experimental details).
Positive controls without drug addition (− Erm) are in the left columns. Right columns: erythromycin added to infected cells (+ Erm). –/–: non-induced bacteria, no addition of rhamnose after macrophage infection; –/+: non-induced bacteria, 10 mM rhamnose added after infection; +/–: pre-induced bacteria, no addition of rhamnose after macrophage infection; +/+: pre-induced bacteria, 10 mM rhamnose added after infection; LF003: constitutive expression of ermC.
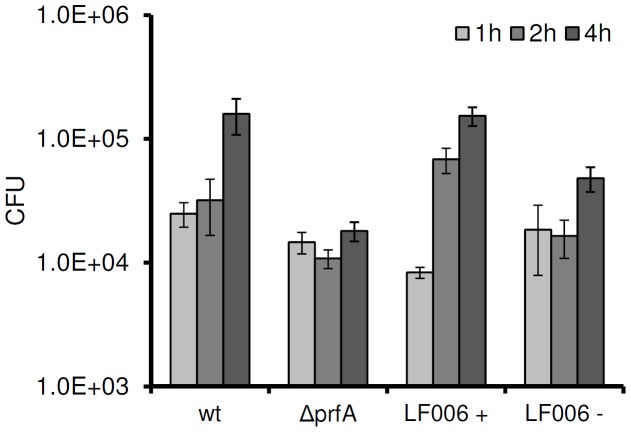
L. monocytogenes strain LF006 features a chromosomal ΔprfA deletion [24] of the gene encoding the regulator PrfA, which is complemented by prfA under Prha control on a single copy pLF6 insertion. When LF006 was pre-induced with 10 mM rhamnose prior to infection of THP1 macrophages, virulence could be fully restored (Fig. 6). Non-induced LF006 did not significantly differ in intracellular multiplication from the negative control (ΔprfA) one and two hours post infection. However, four hours post infection a slight increase in intracellular cell counts was observed (p<0.0001).
Figure 6. Intracellular multiplication of prfA-negative and Prha-dependent trans-complemented L. monocytogenes after infection of human THP1 macrophages.
wt: wild type; ΔprfA: prfA knock out; LF006 +: single copy prfA under control of Prha (pre-induced overnight with 10 mM rhamnose); LF006 −: non-induced.
Discussion
Inducible gene expression systems for bacteria are important tools for biotechnology applications and basic research. Several well defined vectors are available for model organisms such as E. coli or B. subtilis In contrast, very few systems for inducible gene expression in the opportunistic pathogen L. monocytogenes are available, which can be useful for tuned expression of virulence genes or genes encoding potentially toxic products. Dancz and co-workers reported an IPTG-inducible pLIV vector, where a synthetic promoter PSPAC was used in combination with the lacO id operator [20] and lacI, placed under control of the constitutive Piap (P60) promoter. Similar vectors have been developed by others [25]. However, negatively regulated promoters are often leaky, due to less stringent repression and feature poor repression [22]. This renders cloning of potentially lethal or toxic genes difficult or even impossible. Because transcriptional control (i.e., repression) in positively regulated promoters such as Para, Pxyl, or Prha generally is much tighter, our goal was to design such a system for Listeria.
The Prha promoter identified and used in this work is silent in the absence of the sugar, and can be specifically induced in a dose dependent manner by addition of 10–100 µM rhamnose to growing cells. All strains of L. monocytogenes can utilize rhamnose as a single carbon source; additional Prha-binding regulatory proteins are not required. This renders application of Prha simple and straightforward, the promoter may be fused to any gene of interest. In addition, transcription is subject to catabolite repression and the promoter is not active when glucose is present, which provides additional options for Prha-controlled gene expression. However, the exact mechanism of catabolite repression is not clear. It is possible that it is similar to the situation in the maltose and maltodextrin utilization gene clusters, where catabolite expression is independent of the catabolite control protein CcpA, and likely regulated by inducer exclusion [26].
During infection of macrophages, intracellular bacteria featuring Prha-controlled ermC expression were unable to multiply. This can be explained by the fact that rhamnose is not actively transported over the mammalian (macrophage) cytoplasmic membrane [27], which should effectively prevent induction of Prha in intracellular bacteria. Because rhamnose is generally not available in the cytosol of human or animal cells, Prha may be employed to shut down gene expression upon entry of L. monocytogenes into a host cell. Moreover, catabolite repression by intracellular glucose deposits may also contribute to prevent expression from the Prha promoter. This feature can be useful for development of live attenuated vaccine vectors based on the intracellular lifestyle of this pathogen. The opposite approach, e.g., onset of gene expression after entry into the host cytosol has been realized using PactA [28]. This promoter is PrfA-dependent and specifically induced in intracellular L. monocytogenes. The aims were to kill and lyse intracellular L. monocytogenes for use as DNA delivery vehicles into eukaryotic cells, by expression of bacteriophage cell wall hydrolase ply 118 under control of PactA [29].
The challenge in designing L. monocytogenes as a delivery vehicle for the allocation of proteins to the immune system is to attenuate the bacterium in such a way that it retains the ability to invade potential host cells, however, without causing extensive collateral damage. So far, attenuated strains often featured disruption of essential virulence factors, such as actA, inlB or prfA [30], [31], [32], [33]. An alternative strategy is to create L. monocytogenes auxotroph mutants, such as dal and dat null strains defective in D-alanine synthesis [34]. All of these strains have been applied to stimulate specific T-cell responses in vivo. One particular prfA defective strain (Lm-LLO-E7) was used as a vaccine against invasive carcinoma of the cervix in a clinical phase I study.
PrfA is a soluble cytosolic protein that controls transcription of key virulence factors in L. monocytogenes. We tested Prha to trigger prfA expression in an infection model. The prfA-negative phenotype could be fully complemented, but only when expression of the gene in strain LF006 was pre-induced with rhamnose prior to macrophage infection (Fig. 6). In this case, induction of prfA resulted in an overproduction of the protein compared to the wild type, and it is reasonable to assume that the intracellular concentration of PrfA was sufficiently high to ensure its presence and function over the next few generations, i.e., cell divisions. Hence, even when rhamnose as inducer of prfA expression is removed upon invasion and entry into host cells, the protein likely was present at a sufficient threshold concentration to activate transcription of the PrfA regulon in the daughter cells. Further experiments would be required to demonstrate that these cells progressively lose virulence after prolonged incubation, division and intracellular life. In fact, finely tuned prfA induction from Prha by lower rhamnose concentrations could yield the individually desired control of a corresponding phenotype.
In these experiments, we also noted that virulence of non-induced LF006 was not as strictly attenuated as the prfA negative control, which may suggest a low level background expression from the Prha promoter under these specific conditions. It is not unreasonable to assume that even very small amounts of PrfA may be sufficient to slightly elevate intracellular counts of non-induced LF006. Nevertheless, the possibility of employing Prha for attenuation of virulence genes in intracellular L. monocytogenes for application of the organism as live vaccine remains an attractive option.
Materials and Methods
Bacterial Strains and Culture Conditions
L. monocytogenes was cultured in half-strength Brain Heart Infusion (1/2 BHI) broth at 30° or 37°C (macrophage infection model). Chloramphenicol (10 µg/ml) or erythromycin (7.5 µg/ml) was added to the media as needed. Rhamnose or other sugars were used at different concentrations, up to a maximum of 50 mM. To determine the utilization of rhamnose by L. monocytogenes, cells were incubated in Luria-Bertani (LB) broth substituted with 50 mM glucose and 50 mM rhamnose, respectively. Escherichia coli SM10 and XL1-blue MRF’ (Stratagene) were routinely cultured in LB broth at 37°C and with the addition of chloramphenicol (10 µg/ml for XL1-blue MRF’ and 20 µg/ml for SM10) when appropriate. All strains used in this study are summarized in Table 1.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic | Reference or Source |
| E. coli | ||
| XL1-Blue MRF’ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1lac[F′ proABlacI qZ ΔM15 Tn10 (Tetr)] | Stratagene |
| SM10 | λpir | R. Calendar |
| L. monocytogenes | ||
| 10403S | wild type, StmR | D. Portnoy |
| EGDe | wild type | J. Kreft |
| EGDe ΔprfA | prfA deleted | J. Kreft [24] |
| LF001 | 10403S derivative, tRNAArg::Prha gfp | This work |
| LF002 | 10403S derivative, tRNAArg::Prha ermC | This work |
| LF003 | 10403S derivative, tRNAArg::pPL3 | This work |
| LF004 | 10403S derivative, tRNAArg::gfp | This work |
| LF005 | 10403S derivative, tRNAArg::Phyp gfp | This work |
| LF006 | EGDe ΔprfA, tRNAArg::Prha prfA | This work |
| Plasmids | ||
| pPL2 | cat cat; E. coli / L. monocytogenes shuttle vector; thermosensitive ori for Listeria | [34] |
| pPL3e | pPL2 derivative, cat ermC gfp; E. coli / L. monocytogenes shuttle vector;thermosensitive ori for Listeria | D. Higgins [35] |
| pLF1 | pPL2 derivative, Prha | This work |
| pLF2 | pPL2 derivative, Prha gfp | This work |
| pLF3 | pPL2 derivative, Prha ermC | This work |
| pLF4 | pPL2 derivative, gfp | This work |
| pLF5 | pPL2 derivative, Phyp gfp | D. Higgins |
| pLF6 | pPL2 derivative, Prha prfA | This work |
| pLEB599 | Source of ermC | T. Takala |
Bioinformatics
The lmo2846-lmo2850 operon in the L. monocytogenes genome [9] was identified and analyzed in silico (CLC Main Workbench software; CLC Bio, Aarhus, Denmark). Putative -10 and -35 regions were identified by a promoter-finding algorithm (BPROM, http://linux1.softberry.com/berry.phtml).
Cloning Procedures
Plasmid pPL2 was used for cloning and single copy insertion into a tRNAArg gene via bacteriophage PSA site-specific integrase. The plasmid remains stable in the absence of drug selection, and does not cause polar effects [35].
The Prha promoter region was PCR amplified using primers Prha-f (5′- ATTGCGAGCTCTATTCCGTGATAATTTGG-3′) and Prha-r (5′- AAACGGCCGACTCATTTT AGTTAAGCGC-3′), yielding a 671 bp product. Following digestion with SacI and EagI (sites are underlined), the fragment was inserted into pPL2 to yield pLF1. The gfp gene was excised from pPL3 [36] (kindly provided by D. Higgins, Harvard Medical School, USA), using enzymes EagI and KpnI, purified by gel extraction, and inserted into the corresponding sites in pLF1 downstream of Prha. The ermC gene was amplified from pLEB579 (kindly provided by T. Takala, University of Helsinki, Finland), using primers ermC-f (5′-AAACGGCCGACCAAA TTAAAGAGGGTTATAATG-3′) and ermC-r (5′-AAAGGTACCGAAAA ACAAGTTAAGGGATGC-3′), digested with EagI and KpnI (sites are underlined), and cloned into pPL2. Ligation reactions were transformed into E. coli SM10 by electroporation, clones containing the desired inserts were identified by colony PCR, and plasmids recovered by a standard alkaline lysis method. The promoter region and inserts of interest were sequenced. Details of the plasmids and strains are listed in Table 1.
E. coli SM10 carrying pLF-derived plasmids were then used for vector transformation by conjugation into L. monocytogenes 10403S, applying a mating procedure as described previously [35]. L. monocytogenes EGDe was transformed using electroporation [37]. The desired clones were selected on BHI agar containing 200 µg/ml streptomycin and/or 10 µg/ml chloramphenicol, respectively, after incubation for 2–3 days at 30°C.
Fluorescence Reporter Assays
The synthesis of GFP as a reporter for gene expression was monitored using confocal laser scanning microscopy (TCS SPE, Leica, Germany), or a fluorescence plate reader (VICTOR3Multilabel Counter, PerkinElmer, USA). To ensure complete and correct folding of mature GFP proteins, bacteria were harvested from overnight cultures, adjusted to an OD600 nm of 1.0 per ml, and washed 3 times with phosphate buffered saline prior to analysis. For spectrophotometry, 200 µl of the suspension was transferred into a well of a black 96 well plate. Each sample was analyzed in triplicate, and the experiment was independently repeated three times. Arithmetic means and standard deviations are indicated.
Listeria Infection of Macrophages
Human THP1 macrophages [23] were cultured in RPMI 1640 medium supplemented with 20% FBS (Sigma), at 37°C in an atmosphere containing 5% CO2. Approximately 5×105 cells per well of a 24-well plate were seeded, and infected by addition of 1×107 L. monocytogenes strain LF002 or LF006 (Tab. 1), for 1 h (MOI 20). For the infection, rhamnose-induced (10 mM rhamnose) or non-induced control bacteria were taken from overnight cultures incubated at 37°C, adjusted to the desired cell concentration, and washed three times in pre-warmed PBS before use. LF002 infected Macrophages were washed with prewarmed PBS and further incubated in RPMI 1640 medium containing 50 µg/ml gentamycin to inhibit extracellular bacteria. After 1 h of incubation, macrophages were lysed to determine the initial intracellular viable counts (colony forming units, cfu) of L. monocytogenes LF002. Cells were washed three times with prewarmed PBS, scraped off the bottom of each well, and resuspended and lysed by vigorous pipetting and vortexing using ice-cold 0.5% (v/v) Triton X-100. The lysates were diluted and plated for determination of L. monocytogenes cfu. Then, 10 mM rhamnose was added to the remaining infected cells, e.g., (in the remaining wells of the plate), and 7.5 µg/ml erythromycin was added1 h later. These cells were then also lysed using ice-cold 0.5% (v/v) Triton X-100 at different time points. The lysates were diluted and plated for determination of L. monocytogenes cfu from samples taken at 1, 4, 8, and 12 h post infection. Each experiment was performed in triplicate, and independently repeated three times. Means and standard deviations are indicated.
L. monocytogenes strain LF006 was used to complement the ΔprfA mutant from a gene under Prha control (Tab 1). Macrophage infection and determination of intracellular cfu was carried out as described above, using rhamnose pre-induced and non-induced LF006 for infection. Gentamycin (50 µg/ml) was added to inhibit extracellular bacteria after an infection time of 1 h, but rhamnose or any other substance was not added to the infected cells. The cfu of intracellular L. monocytogenes were determined after 1, 2 and 4 h post infection as described above.
Statistical Analyses
Statistical analyses were performed applying ANOVA and student’s t-test algorithms.
Acknowledgments
We thank Darren Higgins (Harvard University), Richard Calendar (University of California, Berkeley), Timo Takala (University of Helsinki), and Juergen Kreft (University of Würzburg) for the gift of plasmids and strains used in this study.
Funding Statement
LF was financially supported by a Feodor-Lynen Research Fellowship of the Alexander von Humboldt Foundation, Bonn, Germany, and JT was enrolled in the Biology Undergraduate Summer School (BUSS) at ETH Zurich and University of Zurich. This study was funded by resources from ETH Zurich. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Freitag NE, Port GC, Miner MD (2009) Listeria monocytogenes - from saprophyte to intracellular pathogen. Nat Rev Microbiol 7: 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cossart P, Toledo-Arana A (2008) Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect 10: 1041–1050. [DOI] [PubMed] [Google Scholar]
- 3. Allerberger F, Wagner M (2010) Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 16: 16–23. [DOI] [PubMed] [Google Scholar]
- 4. Hagens S, Loessner MJ (2007) Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 76: 513–519. [DOI] [PubMed] [Google Scholar]
- 5. Allerberger F (2003) Listeria: growth, phenotypic differentiation and molecular microbiology. FEMS Immunol Med Microbiol 35: 183–189. [DOI] [PubMed] [Google Scholar]
- 6.Graves LM, Helsel LO, Steigerwalt AG, Morey RE, Daneshvar MI, et al.. (2009) Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int J Syst Evol Microbiol. [DOI] [PubMed]
- 7. Wendlinger G, Loessner MJ, Scherer S (1996) Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 142 (Pt 4): 985–992. [DOI] [PubMed] [Google Scholar]
- 8. Xue J, Murrieta CM, Rule DC, Miller KW (2008) Exogenous or L-rhamnose-derived 1,2-propanediol is metabolized via a pduD-dependent pathway in Listeria innocua. Appl Environ Microbiol 74: 7073–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, et al. (2001) Comparative genomics of Listeria species. Science 294: 849–852. [DOI] [PubMed] [Google Scholar]
- 10. Egan SM, Schleif RF (1993) A regulatory cascade in the induction of rhaBAD. J Mol Biol 234: 87–98. [DOI] [PubMed] [Google Scholar]
- 11. Korndorfer IP, Fessner WD, Matthews BW (2000) The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution. J Mol Biol 300: 917–933. [DOI] [PubMed] [Google Scholar]
- 12. Moralejo P, Egan SM, Hidalgo E, Aguilar J (1993) Sequencing and characterization of a gene cluster encoding the enzymes for L-rhamnose metabolism in Escherichia coli. J Bacteriol 175: 5585–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muiry JA, Gunn TC, McDonald TP, Bradley SA, Tate CG, et al. (1993) Proton-linked L-rhamnose transport, and its comparison with L-fucose transport in Enterobacteriaceae. Biochem J 290 (Pt 3): 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhu P, Doan TT, Jeya M, Kang LW, Lee JK (2010) Cloning and characterization of a rhamnose isomerase from Bacillus halodurans. Appl Microbiol Biotechnol. [DOI] [PubMed]
- 15. Yoshida H, Yamaji M, Ishii T, Izumori K, Kamitori S (2010) Catalytic reaction mechanism of Pseudomonas stutzeri L-rhamnose isomerase deduced from X-ray structures. FEBS J 277: 1045–1057. [DOI] [PubMed] [Google Scholar]
- 16. Lee NL, Gielow WO, Wallace RG (1981) Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A 78: 752–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyada CG, Stoltzfus L, Wilcox G (1984) Regulation of the araC gene of Escherichia coli: catabolite repression, autoregulation, and effect on araBAD expression. Proc Natl Acad Sci U S A 81: 4120–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wegerer A, Sun T, Altenbuchner J (2008) Optimization of an E. coli L-rhamnose-inducible expression vector: test of various genetic module combinations. BMC Biotechnol 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cardona ST, Valvano MA (2005) An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54: 219–228. [DOI] [PubMed] [Google Scholar]
- 20. Dancz CE, Haraga A, Portnoy DA, Higgins DE (2002) Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J Bacteriol 184: 5935–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monk IR, Gahan CG, Hill C (2008) Tools for functional postgenomic analysis of listeria monocytogenes. Appl Environ Microbiol 74: 3921–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhavsar AP, Zhao X, Brown ED (2001) Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol 67: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auwerx J (1991) The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia 47: 22–31. [DOI] [PubMed] [Google Scholar]
- 24. Bockmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z (1996) Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol 22: 643–653. [DOI] [PubMed] [Google Scholar]
- 25. Monk IR, Casey PG, Cronin M, Gahan CG, Hill C (2008) Development of multiple strain competitive index assays for Listeria monocytogenes using pIMC; a new site-specific integrative vector. BMC Microbiol 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gopal S, Berg D, Hagen N, Schriefer EM, Stoll R, et al. (2010) Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose. PLoS One 5: e10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brewster DR, Manary MJ, Menzies IS, O’Loughlin EV, Henry RL (1997) Intestinal permeability in kwashiorkor. Arch Dis Child 76: 236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dietrich G, Bubert A, Gentschev I, Sokolovic Z, Simm A, et al. (1998) Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat Biotechnol 16: 181–185. [DOI] [PubMed] [Google Scholar]
- 29. Pilgrim S, Stritzker J, Schoen C, Kolb-Maurer A, Geginat G, et al. (2003) Bactofection of mammalian cells by Listeria monocytogenes: improvement and mechanism of DNA delivery. Gene Ther 10: 2036–2045. [DOI] [PubMed] [Google Scholar]
- 30. Soussi N, Milon G, Colle JH, Mougneau E, Glaichenhaus N, et al. (2000) Listeria monocytogenes as a short-lived delivery system for the induction of type 1 cell-mediated immunity against the p36/LACK antigen of Leishmania major. Infect Immun 68: 1498–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brockstedt DG, Giedlin MA, Leong ML, Bahjat KS, Gao Y, et al. (2004) Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc Natl Acad Sci U S A 101: 13832–13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshimura K, Jain A, Allen HE, Laird LS, Chia CY, et al. (2006) Selective targeting of antitumor immune responses with engineered live-attenuated Listeria monocytogenes. Cancer Res 66: 1096–1104. [DOI] [PubMed] [Google Scholar]
- 33. Shahabi V, Seavey MM, Maciag PC, Rivera S, Wallecha A (2011) Development of a live and highly attenuated Listeria monocytogenes-based vaccine for the treatment of Her2/neu-overexpressing cancers in human. Cancer Gene Ther 18: 53–62. [DOI] [PubMed] [Google Scholar]
- 34. Wallecha A, Maciag PC, Rivera S, Paterson Y, Shahabi V (2009) Construction and characterization of an attenuated Listeria monocytogenes strain for clinical use in cancer immunotherapy. Clin Vaccine Immunol 16: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R (2002) Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 184: 4177–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grundling A, Burrack LS, Bouwer HG, Higgins DE (2004) Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci U S A 101: 12318–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park SF, Stewart GS (1990) High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94: 129–132. [DOI] [PubMed] [Google Scholar]