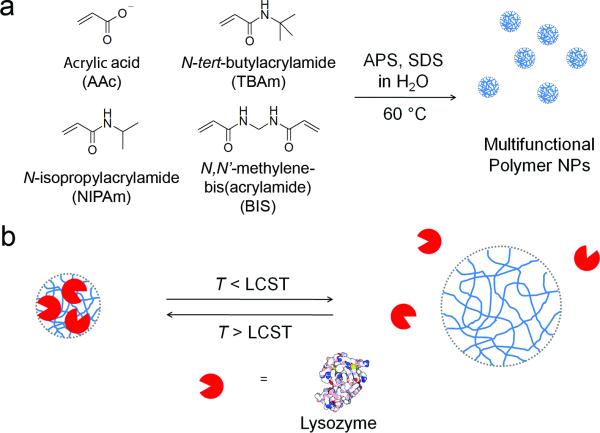
Synthetic nanopartciles (NPs) with an intrinsic affinity for specific proteins are of considerable interest for their potential in biological/biomedical science and biotechnology.[1, 2, 3] In addition to their binding capability, synthetic materials offer the possibility for controlling binding affinity by external stimuli, including light, electromagnetic radiation and temperature;[4] a feature that can be used for remote modulation of capture or release of target proteins in a spatiotemporally controlled manner. In this communication, we report the synthesis and applications of a multifunctional polymer NP with selective protein affinity that can be modulated by external stimuli to “catch-and-release” the target protein.
Synthetic materials that alter their binding affinity to a target molecule in response to an external stimuli have included photoresponsive synthetic receptors for recognition of ions[5] and saccharides,[6] temperature-responsive N-isopropylacrylamide (NIPAm) linear polymers for extraction of atrazine,[7] and temperature-responsive NIPAm-based bulk polymer hydrogels that reversibly change their affinity to target small molecules.[8] More recently, the Lyon and Shea groups reported temperature-responsive NIPAm-based polymer NPs that can change their affinity to doxorubicin,[9] a drug molecule, and melittin, a peptide toxin.[10] However, NPs for protein targets have not been reported. Our goal was to design polymer NPs that possess capability of reversible and temperature-responsive “catch-and-release” of a target protein from a complex biological sample.
There are two separate but not unrelated requirements to create functional materials that incorporate both selective protein affinity and a trigger for the capture and release of the protein cargo. Strong affinity between designed synthetic materials and proteins has been achieved by incorporating complimentary functional groups into synthetic molecular scaffolds,[11] liner polymers,[12] dendritic polymers[13] and synthetic NPs.[2, 3, 10] Based upon our experience with protein and peptide binding to synthetic polymers, we employed an approach to impart protein binding capability to temperature-responsive NIPAm-based NPs by incorporating complimentary functional groups toward a target protein. Lysozyme, a 14 kDa basic protein (pI = 9.3) isolated from chicken egg white, was chosen as the target protein. Acrylic acid (AAc; negatively charged monomer) and N-tert-butylacrylamide (TBAm; hydrophobic monomer) were selected as functional monomers to interact with positively charged and hydrophobic residues of lysozyme. A precipitation polymerization method, previously reported by Kokufuta,[14] Lyon[15] and our group,[3] was employed for synthesis of a small library of multifunctional NPs consisting of various monomer compositions (figure 1a and Table 1). The NPs were synthesized by a thermally initiated free-radical copolymerization of functional monomers and a cross-linking monomer (BIS, 2 mol %) in aqueous solution containing a small amount of sodium dodecyl sulfate (SDS). Following their synthesis, the NPs were purified by dialysis. Dynamic light scattering (DLS) established the NP solutions were mono-disperse with hydrodynamic diameters approximately 80–100 nm (Table 1.).
Figure 1.
(a) Preparation of multifunctional polymer NPs. APS: ammonium persulfate. SDS: sodium dodecyl sulfate (b) Schematic illustration of temperature-responsive “catch-and-release” of a protein by multifunctional polymer NPs. LCST: lower critical solution temperature.
Table 1.
Monomer composition and hydrodynamic diameter of NPs.
| Sample | Monomer content / mol % | Hydrodynamic diameter / nm | ||||
|---|---|---|---|---|---|---|
| AAc | TBAm | NIPAm | BIS | in H2O | in PBS | |
| NP1 | 0 | 40 | 58 | 2 | 80 | N.A.[a] |
| NP2 | 5 | 40 | 53 | 2 | 86 | 96 |
| NP3 | 10 | 40 | 48 | 2 | 89 | 175 |
| NP4 | 5 | 20 | 73 | 2 | 99 | 225 |
Not applicable. Continuous increase of the measured diameter was observed, presumably due to formation of aggregates
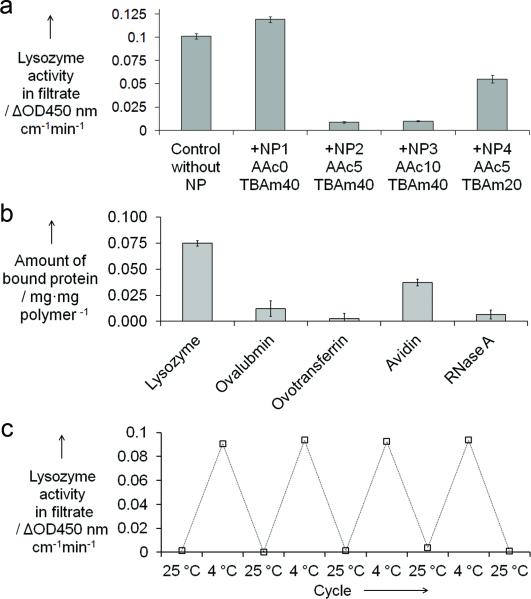
The binding capability of NPs to lysozyme was evaluated by preincubating lysozyme (5 μg/mL) and NPs (800 μg/mL) in phosphate buffered saline (35 mM sodium-phosphate buffer containing 150 mM NaCl, pH 7.3). Solutions were then filtered through a Nanosep membrane filter (MWCO: 100 kDa, Pall Corp.) to filter the NPs and NP-bound lysozyme. The concentration of free lysozyme in the filtrate was estimated by lysozyme activity measurements using a Micrococcus lysodeikticus cell suspension.[16] The results of the binding experiments are summarized in figure 2a. A large decrease in lysozyme activity (> 90%) was observed in the filtrate incubated with NP2 (AAc 5%, TBAm 40%) and NP3 (AAc 10%, TBAm 40%). These results suggest that both hydrophobic and negatively charged groups in the NP contribute to NP-lysozyme capture. We further investigated in situ inhibition capability of NPs by measuring lysozyme activity in presence of 800 μg/mL NP2 and NP3 (Figure S1 in supp. information). This experiment established that 800 μg/mL of NP2 and NP3 inhibited 5 μg/mL of lysozyme. NP2 was particularly effective at inhibiting lysozyme activity to only a few percent of the initial level. Possible explanations include blocking access of the active site of lysozyme by NP2 and/or sequestation of lysozyme into the interior of the polymer network of NP2. Inhibition by NP3 was less pronounced (50% decrease), even though the binding experiment showed that NP3 can capture the majority (> 90%) of lysozyme from solution. In this case, binding to the NP3 apparently does not effectively prevent access of the substrate to the active site of lysozyme. It is interesting to note however, that an increased AAc content does not result in enhanced inhibition of lysozyme activity. This observation implies that the balance between hydrophobic and negatively charged groups in the NP is a critical factor in determining the binding and inhibition capability of the synthetic polymer NPs.
Figure 2.
(a) Depletion of lysozyme (5 μg/mL) from solutions after incubation with NPs (800 μg/mL) at room temp. (b) Binding selectivity of NP2 at room temp. NP2: 2.0 mg/mL. Protein conc: 200 μg/mL. (c) Temperature-responsive “catch-and-release” property of lysozyme (5 μg/mL) by NP2 (2.0 mg/experiments are carried out in PBS (35 mM sodium-phosphate buffer containing 150 mM NaCl, pH 7.3)
Based on these results, NP2 was selected for further study. Lysozyme activity (5 μg/mL) was measured in the presence of various concentrations (0–2.0 mg/mL) of NP2. The 50% inhibition concentration (IC50) of NP2 was determined to be 73 μg/mL, and near complete inhibition is observed in the presence of 1.5 mg/mL of NP2 (Figure S2 in supp. information). Thus a 15- and 300-fold excess of NP2 (in weight) can inhibit half or nearly all, respectively, of lysozyme in solution. The binding selectivity of NP2 was characterized by comparing the uptake of lysozyme and four different proteins. As shown in Figure 2b, 0.075 mg and 0.037 mg of lysozyme and avidin were absorbed per mg of NP2, while only negligible amounts of ovalubmin and ovotransferrin were absorbed by NP2. Under the condition of the experiment (pH = 7.3), lysozyme (pI = 9.3), avidin (pI = 10.0) and RNAse A (pI = 8.6) are positively charged, whereas ovalubmin (pI = 4.7) and ovotransferrin (pI = 6.1) are negatively charged. As NP2 is negatively charged at pH 7.4, the strong interaction between NP2 and the positively charged proteins (lysozyme and avidin) establishes that electrostatic interactions contribute importantly to the interaction. The greater uptake of lysozyme over avidin and RNase A might be explained by the difference in hydrophobicity of those proteins.[17] These results indicate that NP2 has a certain level of binding selectivity that arises from a combination of hydrophobic and complementary electrostatic interactions.
We next investigated the feasibility to release bound lysozyme from the NPs by utilizing the temperature-responsive property of NP2. Below their lowest critical solution temperature (LCST), NIPAm-TBAm copolymers are swollen, highly hydrated and hydrophilic. They are collapsed and more hydrophobic above their LCST due to a entropically driven dissociation of water molecules.[18] NP2 undergoes a volume phase transition from the collapsed “hydrophobic” state to a solvent swollen “hydrophilic” state at approximately 11 °C in PBS (Figure S3 in supporting information).[10, 15] As a consequence, the density of hydrophobic groups and negatively charged groups per unit volume inside the NP decreases. These changes can weaken the NP-protein interactions and trigger release of lysozyme. Figure 2c shows the measured lysozyme activity of the filtrate from each step of the cooling/warming cycles. As can be clearly seen, NP2 was able to “catch-and-release” lysozyme repeatedly in a temperature-responsive manner. Furthermore it was possible to recover lysozyme quantitatively by cooling the samples below the LCST. The process is reversible since the NPs did not show significant loss of binding capability even after 4 cycles. Most importantly, the lysozyme activity did not decrease over the repeated “catch-and-release” cycles indicating that binding to NP2 does not cause denaturation of lysozyme, at least within the time scale of this study. The circular dichroism spectra of lysozyme did not significantly change before and after one cycle (Figure S4 in supporting information), supporting the conclusion that “catch-and-release” does not perturb the structural integrity of lysozyme.
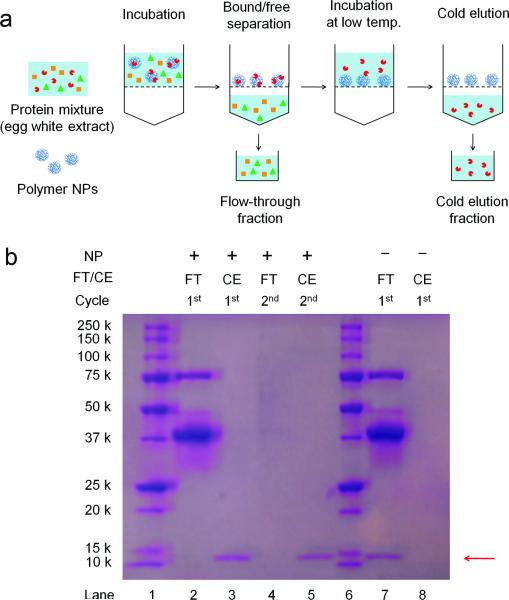
Finally, we evaluated the applicability of NP2 for purification of lysozyme from chicken egg whites. The procedure of “catch-and-release” purification is outlined in Figure 3a. The egg white (diluted 50 times in PBS) and NP2 (2.5 mg/mL) were incubated for 30 min at room temperature. The mixture was then passed through a filter membrane (MWCO: 100 kDa) at room temperature. A control experiment was carried out under identical conditions except for omission of NPs. The proteins in flow-through fractions were analyzed by SDS-PAGE (Figure 3b, lane 2 and 7). While ovalbumin (45 kDa), ovotransferrin (77 kDa) and ovomucoid (28 kDa) could be detected in both filtrates, lysozyme (14 kDa) was only detected in the filtrate of the control experiment. This indicates that NP2 is capable of selectively capturing lysozyme over other major egg white proteins. Subsequently, the same membrane filter unit was filled with a fresh PBS solution and incubated for 30 min at 1 °C to elute lysozyme from the NPs retained on the membrane. Lysozyme was detected in the cold elution fraction from the NP2, none was detected in the control (Figure 3b, lane 3 and 8). Although a trace amount of ovalbumin was also detected in the cold elution fraction from NP2, the purity of the sample could be easily improved after an extra cycle of the “catch-and-release” purification (Figure 3b, lane 5). The amount of recovered lysozyme (based on enzymatic activity) after the first and second cycles was estimated to be 84% and 79% respectively. The specific activity of purified lysozyme was 41579 units/mg, which was comparable to that of a commercially avalible product.
Figure 3.
(a) Schematic of “catch-and-release” lysozyme purification from egg whites. (b) SDS-PAGE analysis of flow-through fraction (FT) and cold elution fraction (CE) after Coomassie Brilliant Blue R-250 staining. Lane 1 and 6: molecular weight markers. Lane 2: FT from 1st cycle. Lane 3: CE from 1st cycle. Lane 4: FT from 2nd cycle. Lane 5: CE fraction from 2nd cycle. A control experiment was carried out in absence of NPs. Lane 7: FT from the control. Lane 8: CE from the control.
In conclusion, we have demonstrated the potential for multifunctional polymer NPs as temperature-responsive “catch-and-release” machines for sequestering target proteins. NIPAm based NPs with various combinations of hydrophobic and negatively charged monomers were screened for in situ lysozyme affinity and inhibition capability. NPs with optimized composition selectively captured lysozyme without denaturation. The protein could be subsequently released from the NP by simply cooling to 1 °C. These results suggest that designed multifunctional NPs have the potential for use as tools for the spatiotemporally controlled capture and release of proteins. This property and the NPs inherent selectivity was successfully applied for “catch-and-release” purification of lysozyme from chicken egg whites. Since NPs with similar composition can selectively capture or release target peptides10 and proteins in a spatiotemporally controlled manner, we envision this concept can find utility in a number of areas in biological/biomedical science and biotechnology.
Supplementary Material
Acknowledgments
Financial support from the National Institutes of Health (GM 080506) is gratefully acknowledged. B.K.L. was a recipient of Allergan Undergraduate Research Fellowship.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- [1].Duncan R. Nat. rev. Drug discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]; Cedervall T, Lynch I, Foy M, Berggard T, Donnelly SC, Cagney G, Linse S, Dawson KA. Angew. Chem. Int. Ed. Engl. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]; Linse S, Cabaleiro-Lago C, Xue WF, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hoshino Y, Koide H, Urakami T, Kanazawa H, Kodama T, Oku N, Shea KJ. J. Am. Chem. Soc. 2010;132:6644–6645. doi: 10.1021/ja102148f. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hoshino Y, Shea KJ. J. Mater. Chem. 2011;21:3517–3521. [Google Scholar]
- [2].Rotello VM, Fischer NO, McIntosh CM, Simard JM. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5018–5023. doi: 10.1073/pnas.082644099. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rotello VM, You CC, Verma A. Soft Matter. 2006;2:190–204. doi: 10.1039/b517354j. [DOI] [PubMed] [Google Scholar]
- [3].Hoshino Y, Urakami T, Kodama T, Koide H, Oku N, Okahata Y, Shea KJ. Small. 2009;5:1562–1568. doi: 10.1002/smll.200900186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stuart MAC, Huck WTS, Genzer J, Muller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, Winnik F, Zauscher S, Luzinov I, Minko S. Nat. Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- [5].Shinkai S, Ogawa T, Nakaji T, Kusano Y, Nanabe O. Tetrahedron Lett. 1979:4569–4572. [Google Scholar]; Shinkai S, Nakaji T, Nishida Y, Ogawa T, Manabe O. J. Am. Chem. Soc. 1980;102:5860–5865. [Google Scholar]; Shinkai S, Nakaji T, Ogawa T, Shigematsu K, Manabe O. J. Am. Chem. Soc. 1981;103:111–115. [Google Scholar]
- [6].Shinmori H, Takeuchi M, Shinkai S. J. Chem. Soc. Perk. Trans. 2. 1996:1–3. [Google Scholar]; Shinkai S, Shinmori H, Takeuchi M. J. Chem. Soc. Perk. Trans. 2. 1998:847–852. [Google Scholar]
- [7].Gonzalez SO, Furyk S, Li CM, Tichy SE, Bergbreiter DE, Simanek EE. J. Polym. Sci. Part. A. Pol. Chem. 2004;42:6309–6317. [Google Scholar]
- [8].Oya T, Enoki T, Grosberg AY, Masamune S, Sakiyama T, Takeoka Y, Tanaka K, Wang GQ, Yilmaz Y, Feld MS, Dasari R, Tanaka T. Science. 1999;286:1543–1545. doi: 10.1126/science.286.5444.1543. [DOI] [PubMed] [Google Scholar]
- [9].Lyon LA, Serpe MJ, Yarmey KA, Nolan CM. Biomacromolecules. 2005;6:408–413. doi: 10.1021/bm049455x. [DOI] [PubMed] [Google Scholar]
- [10].Hoshino Y, Haberaecker WW, III, Kodama T, Zeng Z, Okahata Y, Shea KJ. J. Am. Chem. Soc. 2010;132:13648–13650. doi: 10.1021/ja1058982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hamilton AD, Yin H. Angew. Chem. Int. Ed. Engl. 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- [12].Schrader T, Koch SJ, Renner C, Xie XL. Angew. Chem. Int. Ed. Engl. 2006;45:6352–6355. doi: 10.1002/anie.200601161. [DOI] [PubMed] [Google Scholar]
- [13].Haag R, Dernedde J, Rausch A, Weinhart M, Enders S, Tauber R, Licha K, Schirner M, Zugel U, von Bonin A. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19679–19684. doi: 10.1073/pnas.1003103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kokufuta E, Ito S, Ogawa K, Suzuki H, Wang BL, Yoshida R. Langmuir. 1999;15:4289–4294. [Google Scholar]
- [15].Lyon LA, Debord JD. Langmuir. 2003;19:7662–7664. [Google Scholar]
- [16].Shugar D. Biochim. biophys. acta. 1952;8:302–309. doi: 10.1016/0006-3002(52)90045-0. [DOI] [PubMed] [Google Scholar]
- [17].Durance TD, Nakai S. J. Food. Sci. 1988;53:1096–1102. [Google Scholar]; Ghose S, McNerney TM, Hubbard B. Biotechnol. Bioeng. 2004;87:413–423. doi: 10.1002/bit.20125. [DOI] [PubMed] [Google Scholar]
- [18].Schild HG. Prog. Polym. Sci. 1992;17:163–249. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.